Multiple sequence alignment Jarno Tuimala Scoring matrices Uses










- Slides: 27

Multiple sequence alignment Jarno Tuimala

Scoring matrices

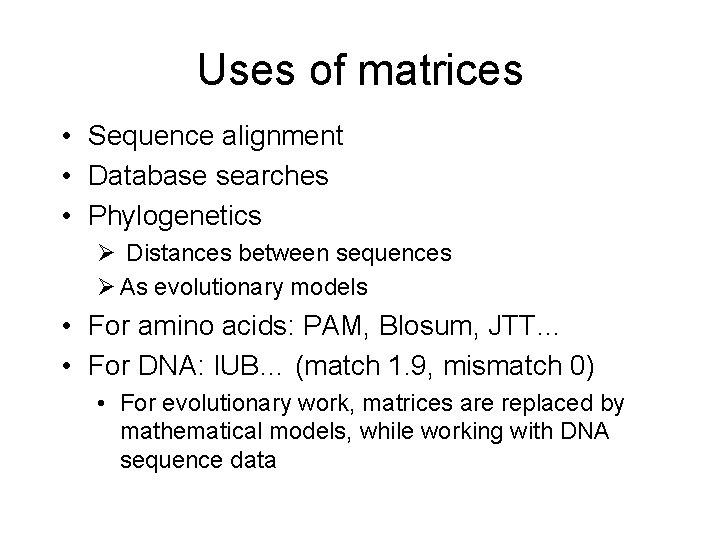
Uses of matrices • Sequence alignment • Database searches • Phylogenetics Ø Distances between sequences Ø As evolutionary models • For amino acids: PAM, Blosum, JTT… • For DNA: IUB… (match 1. 9, mismatch 0) • For evolutionary work, matrices are replaced by mathematical models, while working with DNA sequence data

Muunnettu kuvista: http: //www. bigchalk. com/cgi-bin/Web. Objects/WOPortal. woa/wa/HWCDA/file? fileid=18373&flt=ga Adeniini Guaniini Sytosiini Tymiini

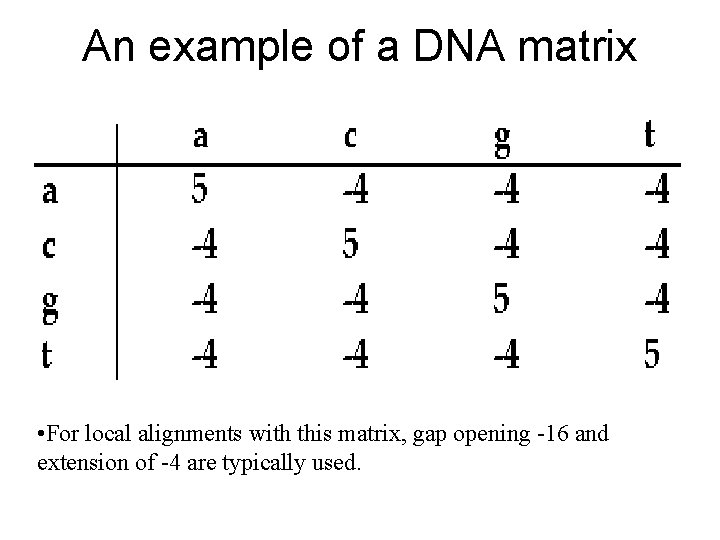
An example of a DNA matrix • For local alignments with this matrix, gap opening -16 and extension of -4 are typically used.

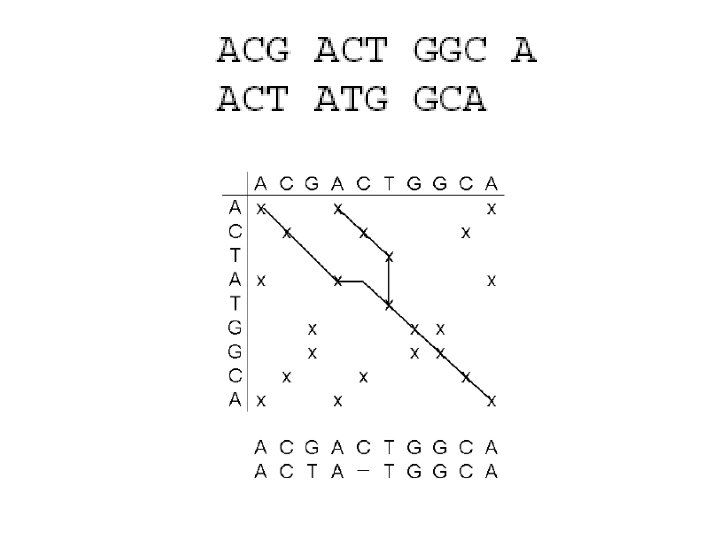
Sequence alignment

How to align sequences • On paper / with computer – Description of alignment for computer: • scoring matrix • gap penalties • Aligning is not objective – Check the results computer gives you! • Alignments can be used for – searching conserved sequence areas – searching point mutations – studying evolution of genes and species

Gap penalties • Gap are evolutionarily expensive. – Opening is more costly than extension – Affine gap model • Mathematically – P = c + gd – P is the total gap penalty – c is gap opening penalty – d is extension penalty – g is the (lenght of the gap - 1)


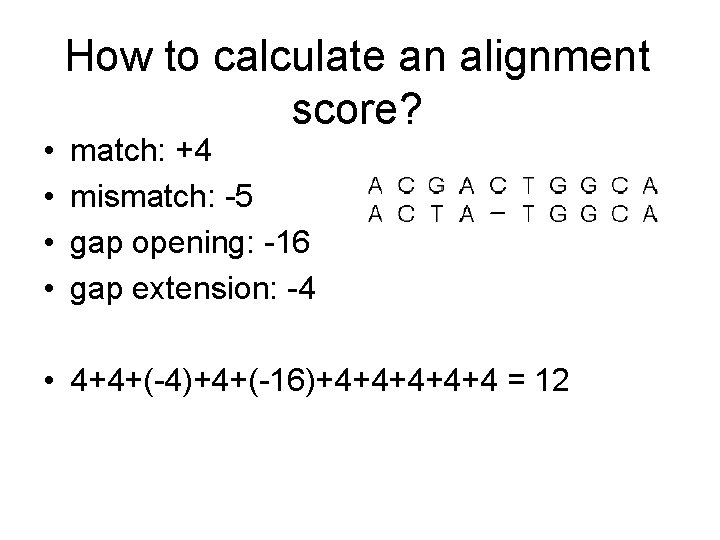
• • How to calculate an alignment score? match: +4 mismatch: -5 gap opening: -16 gap extension: -4 • 4+4+(-4)+4+(-16)+4+4+4 = 12

Multiple sequence alignment (MSA)

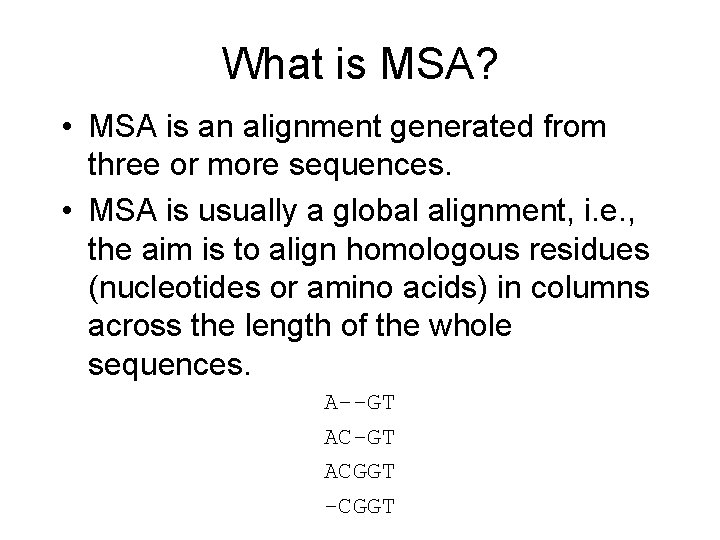
What is MSA? • MSA is an alignment generated from three or more sequences. • MSA is usually a global alignment, i. e. , the aim is to align homologous residues (nucleotides or amino acids) in columns across the length of the whole sequences. A--GT ACGGT -CGGT

Alignability of sequences • If the similarity of sequences drops too low, sequences can’t be reliably aligned (accuracy drops below acceptable). – For proteins <20% similarity – For DNA <~75% similarity • This cut-off is called twilight zone. • In other words, twilight zone marks the sequence similarity below which the observed similarity is mainly due to random variation, and not due to evolution.

MSA and dynamic programming • There are methods that can produce the optimal alignment (in terms of gap penalties and scoring matrices), but they are computationally very heavy. – Program MSA uses dynamic programming • In practise, dynamic programming would be good for up to about 10 sequences, and is not usually used for MSA. – But for pairwise alignment it can be used.

MSA methods • There are two popular methods to perform a multiple sequence alignment: – Progressive alignment • Clustal (Clustal. W and Clustal. X), Pileup… • Clustal is the most commonly used alignment program – Iterative alignment • SAGA… • We will review the Pileup method first

Progressive alignment

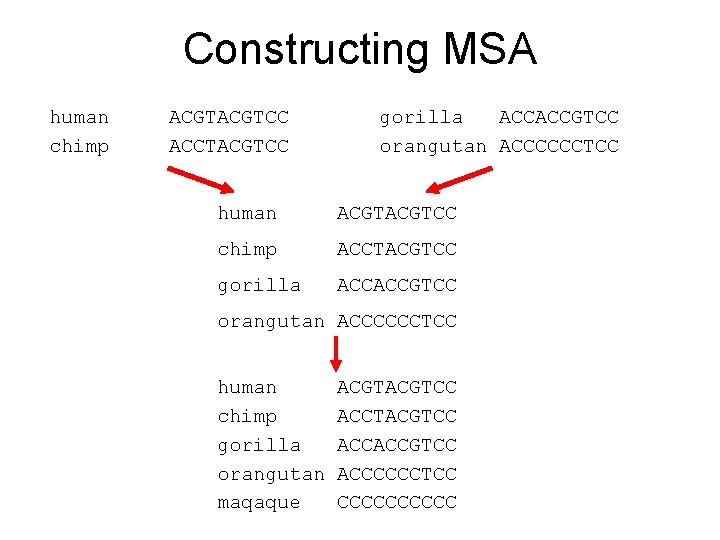
Progressive alignment • Produce pairwise alignment between all the sequences you want to align with MSA. – Dynamic programming, ktup-methods, dot matrix method…(you choose it) • Produce a “guide tree” on the basis of the pairwise distances calculated from pairwise alignments. – UPGMA, neighbor joining (you choose it) • Produce an MSA using the guide tree. – Sequences are aligned in the same order as the guide tree instructs.

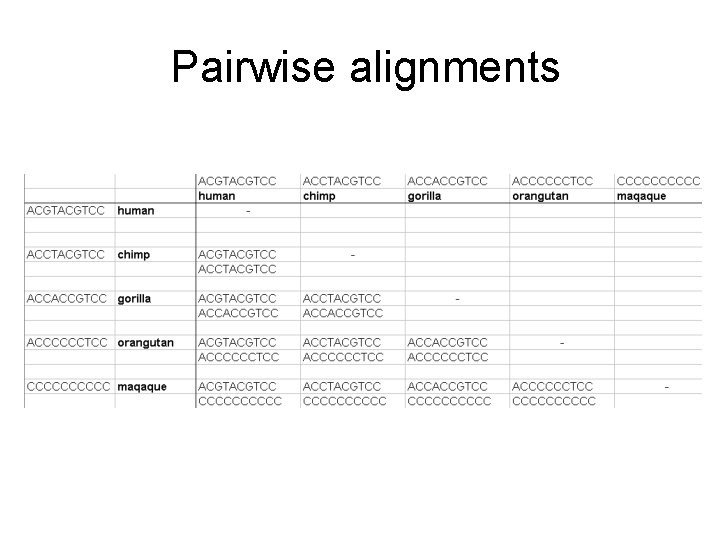
Pairwise alignments

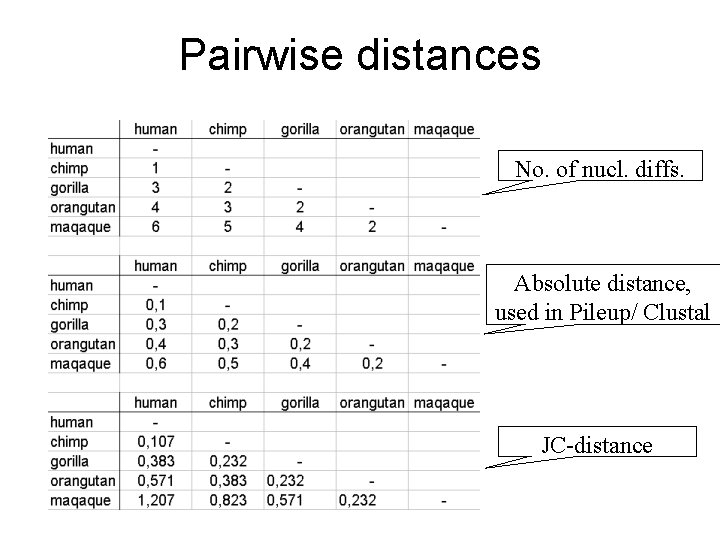
Pairwise distances No. of nucl. diffs. Absolute distance, used in Pileup/ Clustal JC-distance

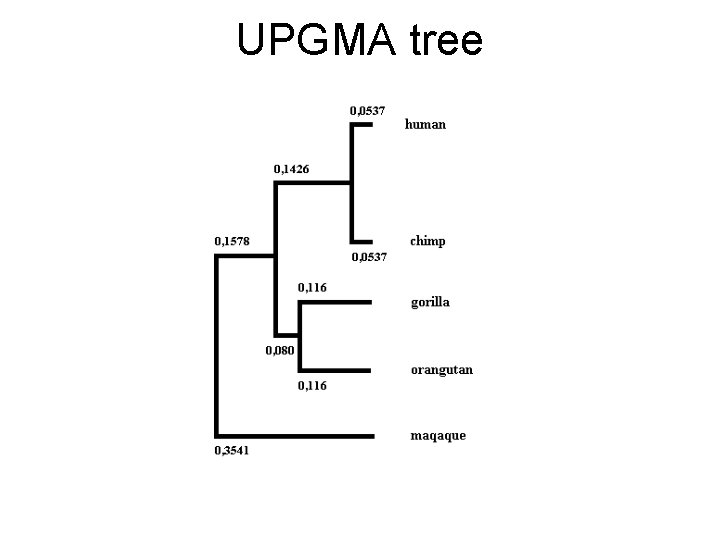
UPGMA • Unweighted Pair Group Method with Arithmetic mean • One of the fastest and tree construction methods • Used in Pileup (GCG package) • Clustal uses neighbor joining, but calculating NJ tree is much more demanding; thus, UPGMA is demonstrated here

UPGMA tree

Constructing MSA human chimp ACGTCC ACCTACGTCC gorilla ACCACCGTCC orangutan ACCCCCCTCC human ACGTCC chimp ACCTACGTCC gorilla ACCACCGTCC orangutan ACCCCCCTCC human chimp gorilla orangutan maqaque ACGTCC ACCTACGTCC ACCACCGTCC ACCCCCCTCC CCCCC

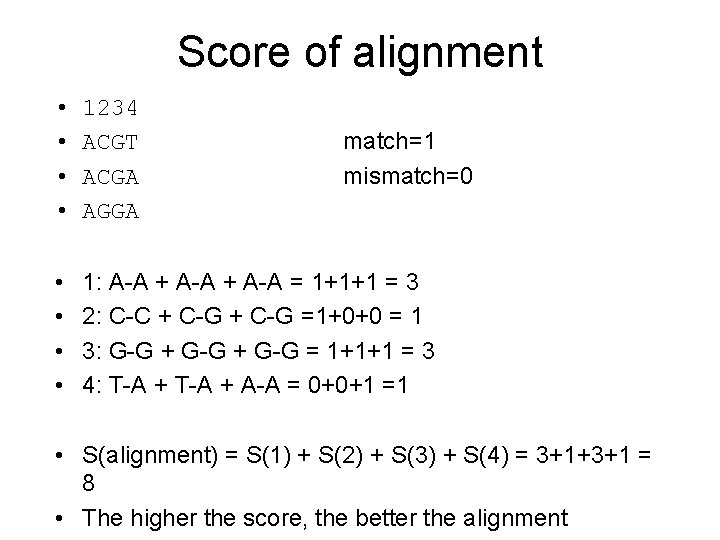
Score of alignment • • 1234 ACGT ACGA AGGA • • 1: A-A + A-A = 1+1+1 = 3 2: C-C + C-G =1+0+0 = 1 3: G-G + G-G = 1+1+1 = 3 4: T-A + A-A = 0+0+1 =1 match=1 mismatch=0 • S(alignment) = S(1) + S(2) + S(3) + S(4) = 3+1+3+1 = 8 • The higher the score, the better the alignment

Progressive alignment - pros and cons • Pros – Fast – Quite accurate • Cons – Once gaps are opened they can never be closed • Errors in the alignment of the first few sequences can have catastrophic effects on the whole alignment

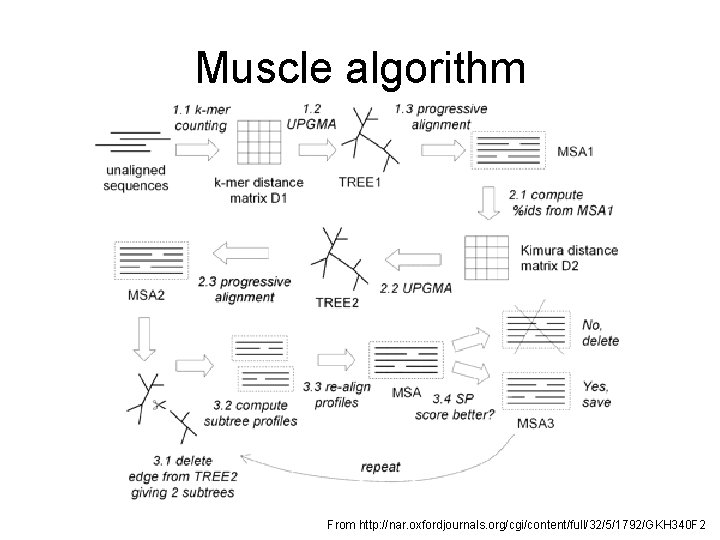
Muscle – both progressive and iterative

Muscle algorithm From http: //nar. oxfordjournals. org/cgi/content/full/32/5/1792/GKH 340 F 2

Muscle – comparison results • As fast as Clustal, but at the same time: • As accurate as T-COFFEE! – T-COFFEE was previously the most accurate alignment method (or software) available