Multiple Sequence Alignment Multiple Alignment versus Pairwise Alignment








- Slides: 22

Multiple Sequence Alignment

Multiple Alignment versus Pairwise Alignment • Up until now we have only tried to align two sequences. • What about more than two? And what for? • A faint similarity between two sequences becomes significant if present in many • Multiple alignments can reveal subtle similarities that pairwise alignments do not reveal

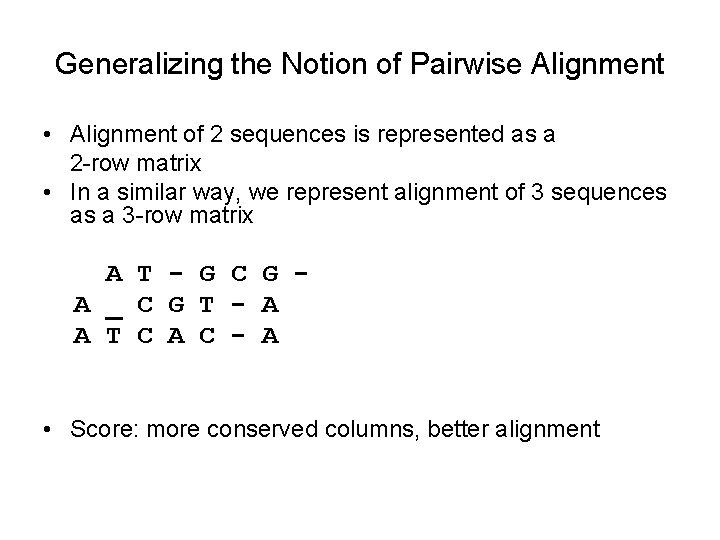
Generalizing the Notion of Pairwise Alignment • Alignment of 2 sequences is represented as a 2 -row matrix • In a similar way, we represent alignment of 3 sequences as a 3 -row matrix A T - G C G A _ C G T - A A T C A C - A • Score: more conserved columns, better alignment

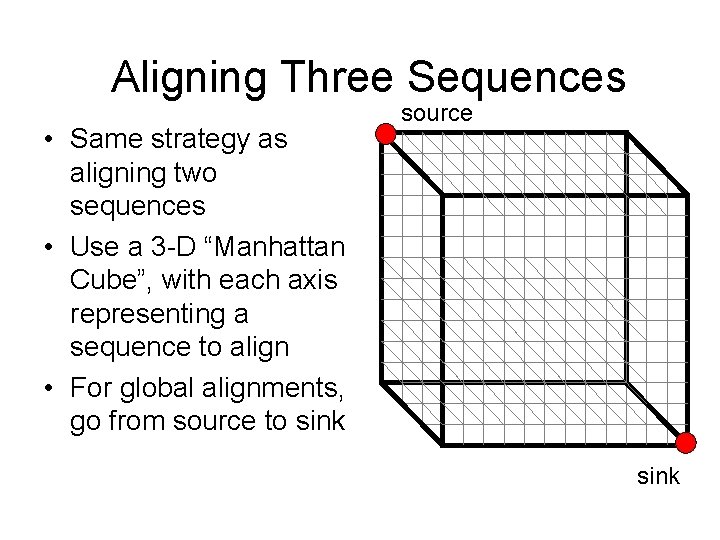
Aligning Three Sequences • Same strategy as aligning two sequences • Use a 3 -D “Manhattan Cube”, with each axis representing a sequence to align • For global alignments, go from source to sink source sink

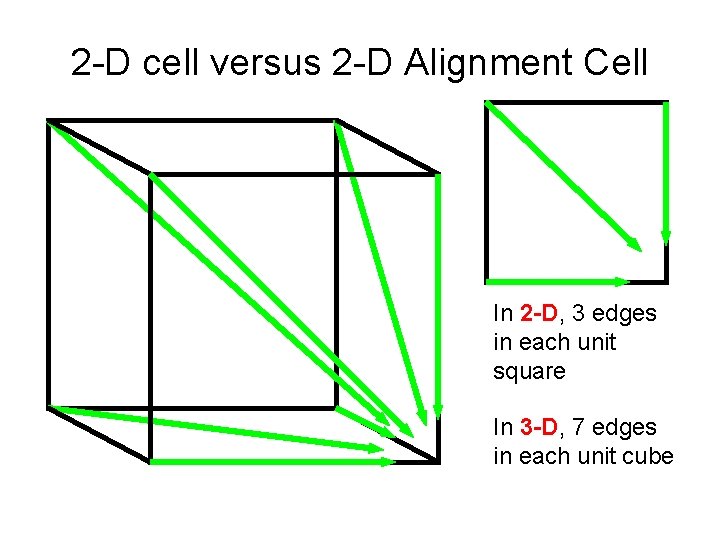
2 -D cell versus 2 -D Alignment Cell In 2 -D, 3 edges in each unit square In 3 -D, 7 edges in each unit cube

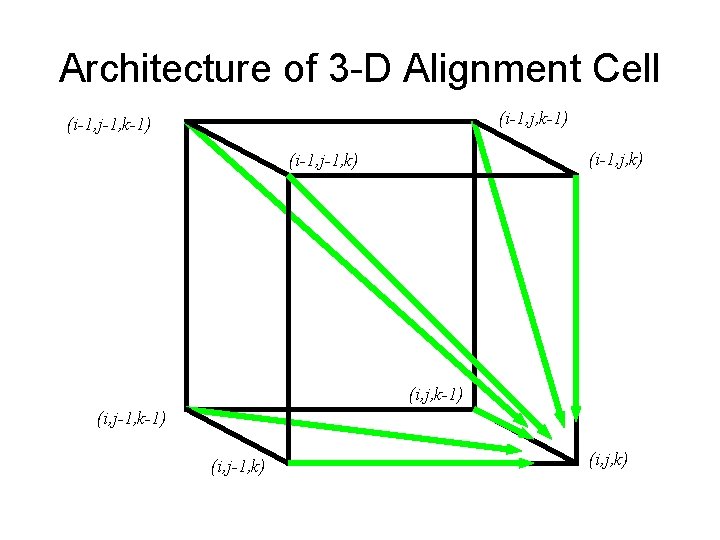
Architecture of 3 -D Alignment Cell (i-1, j, k-1) (i-1, j-1, k-1) (i-1, j, k) (i-1, j-1, k) (i, j, k-1) (i, j-1, k) (i, j, k)

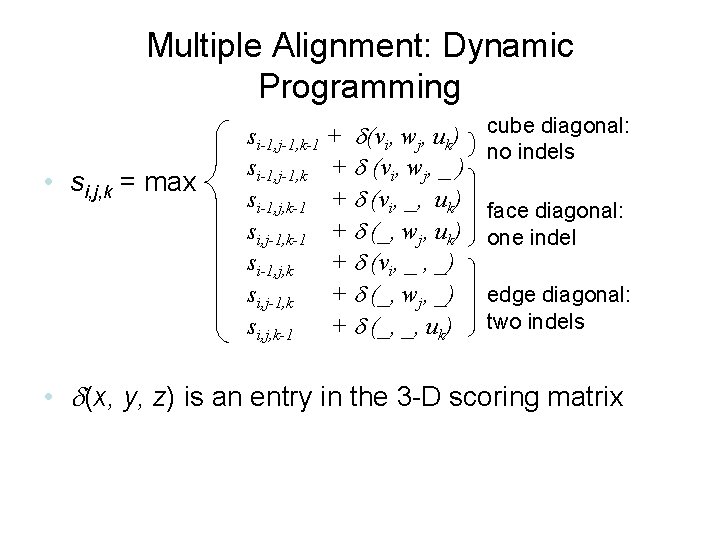
Multiple Alignment: Dynamic Programming • si, j, k = max si-1, j-1, k-1 + (vi, wj, uk) si-1, j-1, k + (vi, wj, _ ) si-1, j, k-1 + (vi, _, uk) si, j-1, k-1 + (_, wj, uk) si-1, j, k + (vi, _ , _) si, j-1, k + (_, wj, _) si, j, k-1 + (_, _, uk) cube diagonal: no indels face diagonal: one indel edge diagonal: two indels • (x, y, z) is an entry in the 3 -D scoring matrix

Multiple Alignment: Running Time • For 3 sequences of length n, the run time is 7 n 3; O(n 3) • For k sequences, build a k-dimensional Manhattan, with run time (2 k-1)(nk); O(2 knk) • Conclusion: dynamic programming approach for alignment between two sequences is easily extended to k sequences but it is impractical due to exponential running time

Practically speaking • Multiple alignment is a hard problem • Yet, it is of extreme practical importance – Comparing several species is a very common task – Doing this for the entire genome is an increasingly common demand! • Several heuristic-based algorithms have been developed that employ greedy, divide-andconquer, dynamic programming approaches in various combinations • The algorithms we will see today are actually used by current multiple aligners

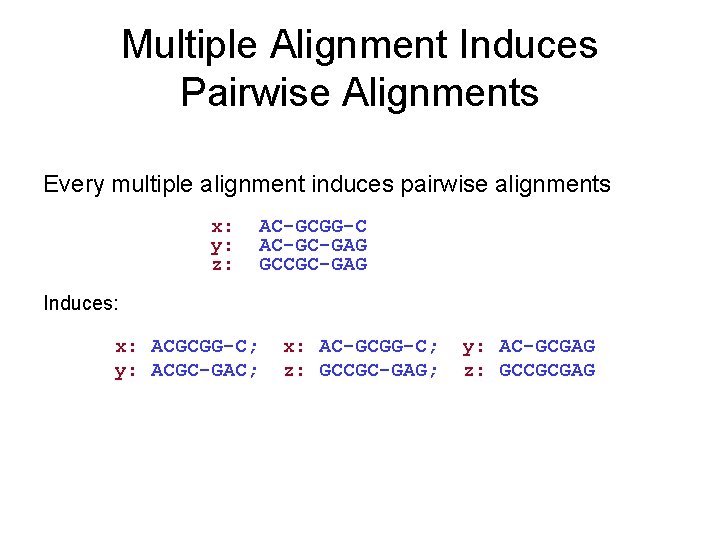
Multiple Alignment Induces Pairwise Alignments Every multiple alignment induces pairwise alignments x: y: z: AC-GCGG-C AC-GC-GAG GCCGC-GAG Induces: x: ACGCGG-C; y: ACGC-GAC; x: AC-GCGG-C; z: GCCGC-GAG; y: AC-GCGAG z: GCCGCGAG

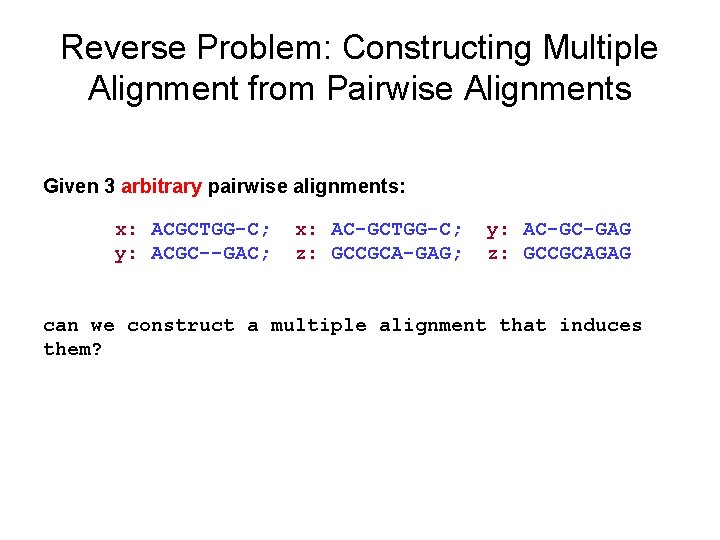
Reverse Problem: Constructing Multiple Alignment from Pairwise Alignments Given 3 arbitrary pairwise alignments: x: ACGCTGG-C; y: ACGC--GAC; x: AC-GCTGG-C; z: GCCGCA-GAG; y: AC-GC-GAG z: GCCGCAGAG can we construct a multiple alignment that induces them?

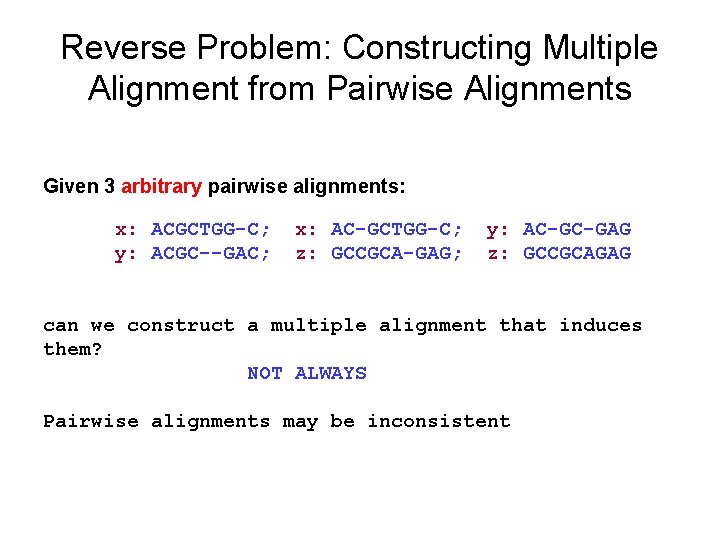
Reverse Problem: Constructing Multiple Alignment from Pairwise Alignments Given 3 arbitrary pairwise alignments: x: ACGCTGG-C; y: ACGC--GAC; x: AC-GCTGG-C; z: GCCGCA-GAG; y: AC-GC-GAG z: GCCGCAGAG can we construct a multiple alignment that induces them? NOT ALWAYS Pairwise alignments may be inconsistent

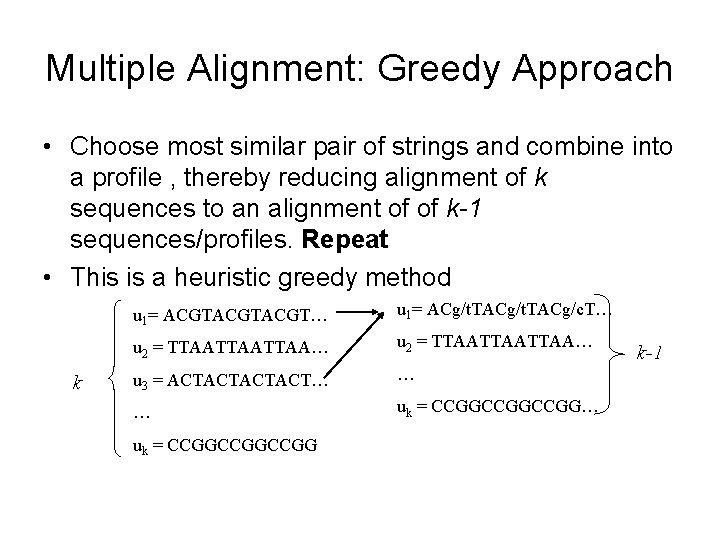
Multiple Alignment: Greedy Approach • Choose most similar pair of strings and combine into a profile , thereby reducing alignment of k sequences to an alignment of of k-1 sequences/profiles. Repeat • This is a heuristic greedy method k u 1= ACGTACGT… u 1= ACg/t. TACg/c. T… u 2 = TTAATTAATTAA… u 3 = ACTACT… … … uk = CCGGCCGGCCGG k-1

Progressive Alignment • Progressive alignment is a variation of greedy algorithm with a somewhat more intelligent strategy for choosing the order of alignments.

Clustal • Popular multiple alignment tool today • Uses “progressive alignment” • Three-step process 1. ) Construct pairwise alignments 2. ) Build Guide Tree 3. ) Progressive Alignment guided by the tree

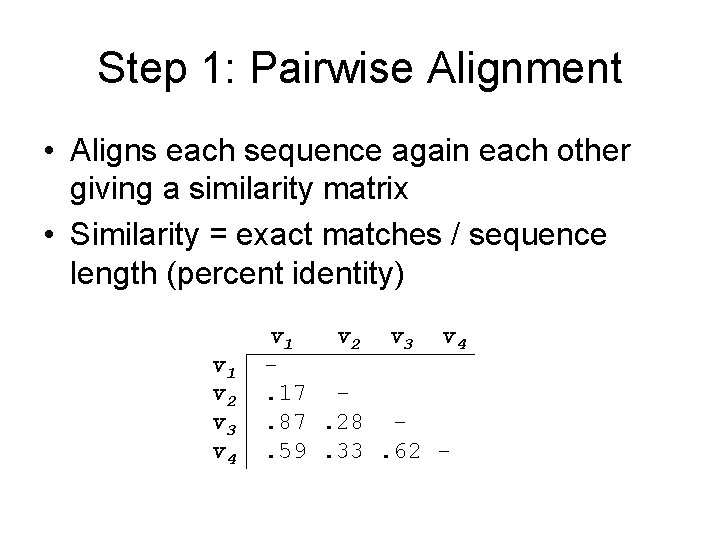
Step 1: Pairwise Alignment • Aligns each sequence again each other giving a similarity matrix • Similarity = exact matches / sequence length (percent identity) v 1 v 2 v 3 v 4. 17. 87. 28. 59. 33. 62 -

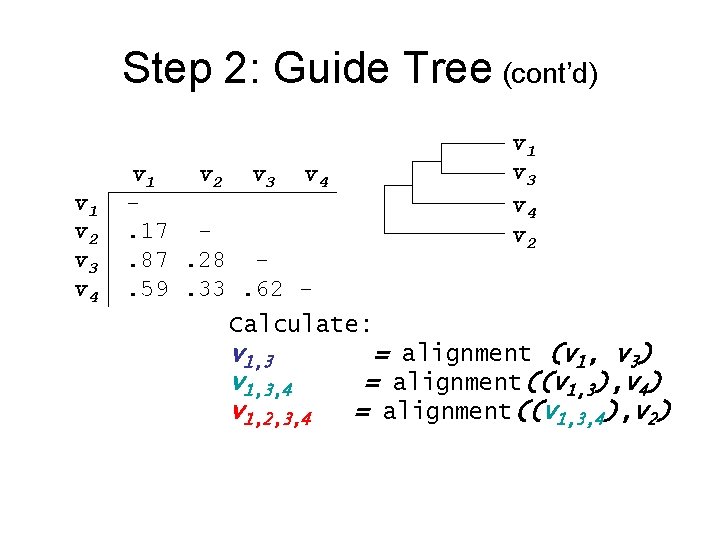
Step 2: Guide Tree • Create Guide Tree using the similarity matrix • Clustal. W uses the “neighbor-joining method” • Guide tree roughly reflects evolutionary relations

Step 2: Guide Tree (cont’d) v 1 v 2 v 3 v 4 v 1 v 3 v 4 v 2 v 1 v 2 v 3 v 4. 17. 87. 28. 59. 33. 62 Calculate: v 1, 3, 4 v 1, 2, 3, 4 = alignment (v 1, v 3) = alignment((v 1, 3), v 4) = alignment((v 1, 3, 4), v 2)

Step 3: Progressive Alignment • Start by aligning the two most similar sequences • Following the guide tree, add in the next sequences, aligning to the existing alignment • An alignment is stored as a “consensus sequence”, to be aligned with other sequences or alignments later • Consensus sequence: Residue a if 75% of aligned sequences have an a at that position. Otherwise “X”.

Evaluation • Evaluating alignment programs is very difficult • What is a benchmark here ? • We haven’t witnessed the process of evolution, so we cannot say for certain what the true alignment of “extant” sequences should be • One approach: “simulate” evolution

Simulating evolution • Generate a random sequence and introduce realistic evolutionary changes to it, along branches of an assumed phylogeny • Substitutions, insertions, deletions, insertion & deletion rates, duplications, introduction of repeat elements, etc.

Evaluating alignment • Once simulation done, take all the sequences at the leaf nodes of the phylogeny (started with root) • Align these sequences using software • Compare computed alignment and known (“true”) alignment – sensitivity and specificity