Multiple Sequence Alignment Clustal W TCoffee Ka Ks











- Slides: 32

Multiple Sequence Alignment Clustal. W TCoffee Ka, Ks, and Ka/Ks Anchored alignment 1

Clustal. W Ø http: //www. ebi. ac. uk/clustalw/ 2

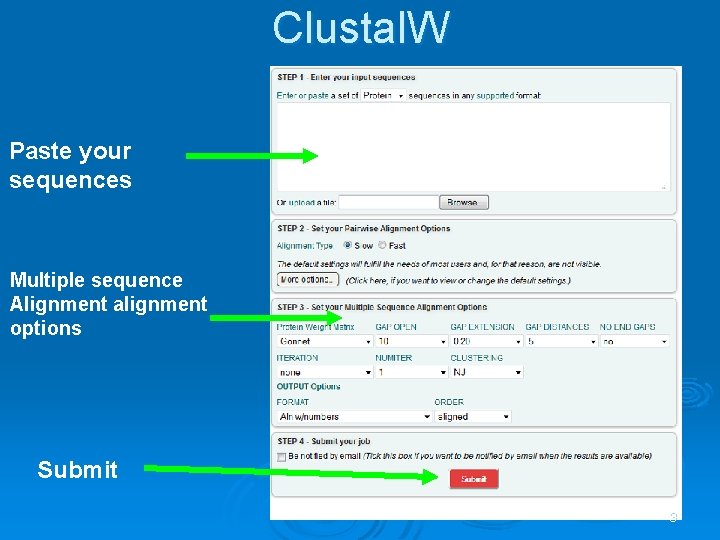
Clustal. W Paste your sequences Multiple sequence Alignment alignment options Submit 3

Exercise Ø Homolo. Gene is a system for automated detection of homologs among annotated genes of several completely sequenced eukaryotic genomes. Ø Download the FASTA sequences of Homolo. Gene: 5276 and align them with Clustal. W 4

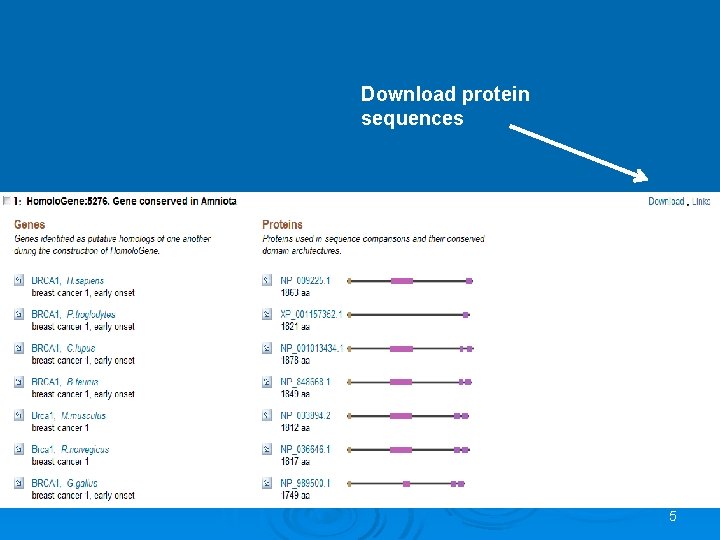
Download protein sequences 5

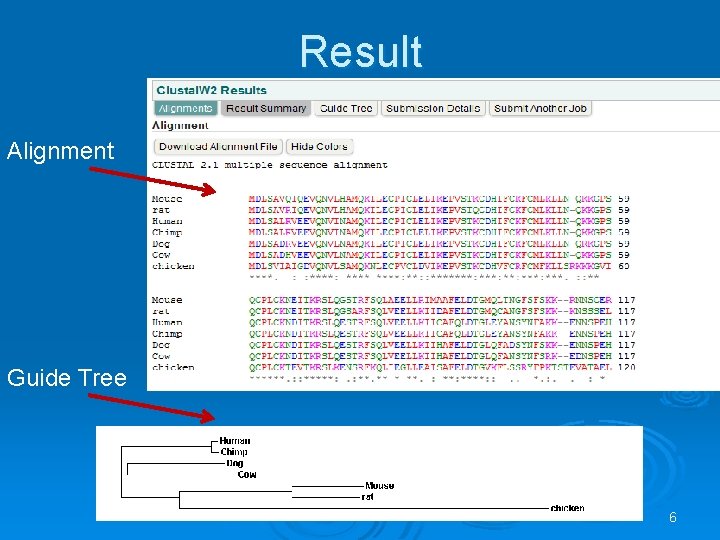
Result Alignment Guide Tree 6

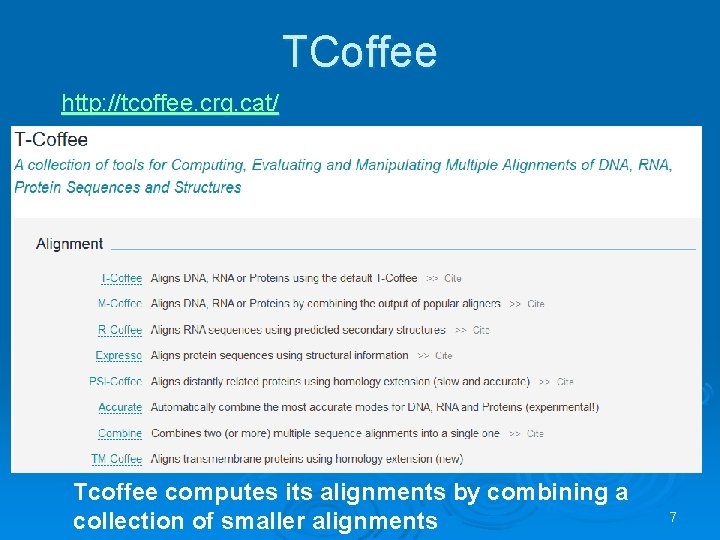
TCoffee http: //tcoffee. crg. cat/ Tcoffee computes its alignments by combining a collection of smaller alignments 7

Alignment at the DNA level based on an alignment at the Protein Level Ø The 18 -k. Da protein plays an important role in fertilization of several abalone species Ø Build a multiple sequence alignment using the following sequences 8

Sequences >gi|604533|gb|AAC 37231. 1| fertilization protein MRSLVLLCVLLMAICAADKKTSVSKENEAAMKVAMMKFLDMKAGVFKEIIEDMGYPITPPQWTTLLYYNR ERLIEFCRSFLALSKKIILLGGNKLNKANFARMGRILGWKSQWAVRQRQWGMVRVSRRHTSTAIAKRIVA MKVADLPCN >gi|604531|gb|AAC 37233. 1| fertilization protein MRFLLLLCVLMGAVSQAVCRKRPNVWGKIVVKEKNKAAMKIGFMEYLDAKLVKFKRHWLVGANWKLQKFE TDEMRYLAIKRLIKVCHGYTIWSQRLIMLKYRPLNEKYFKKVGRYLAWRNYLIVFRMWIGVLKKNLKRSE ITKPMQKLLDTKDGELPCPVRKIHG >gi|604529|gb|AAC 37232. 1| fertilization protein MRSLVLLCVLMAVGCVAFDDVVVSRQEQSYVQRGMVNFLDEEMHKLVKRFRDMRWNLGPGFVFLLKKVNR ERMMRYCMDYARYSKKILQLKHLPVNKKTLTKMGRFVGYRNYGVIRELYADVFRDVQGFRGPKMTAAMRK YSSKDPGTFPCKNEKRRG >gi|604527|gb|AAC 37230. 1| fertilization protein MRSLVLLCVLLMAICAADKKTTVSKENAAAMKIAMIKFLDARAGKFKKRVENMGYPITPPQWTTLLYYNR QRLMEWCHTYVEFSKKIILMGGNKLNKKNFTRMGRIIGWKNQWVLKRRQWEMVRVMRRYKSTAIAKKIVA MKVADLPCN >gi|604525|gb|AAC 37229. 1| fertilization protein MRSLVLLCVLLMAICAADKKSTVSKENAAAMKVAMIKFLDSRTDRFKKRIEKIGYPITPPQYTTLLYYNR ERLMDWCHNYVEVSKKIILLGGNKLNKKNFARMGRIIGWKNQWILKRRQWHMVRVMRRYKASAIAKKIVA MKVADLPCN 9

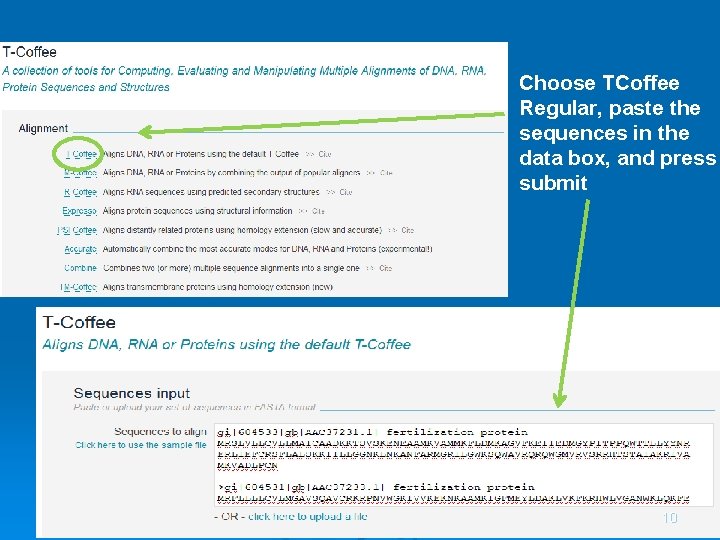
Choose TCoffee Regular, paste the sequences in the data box, and press submit 10

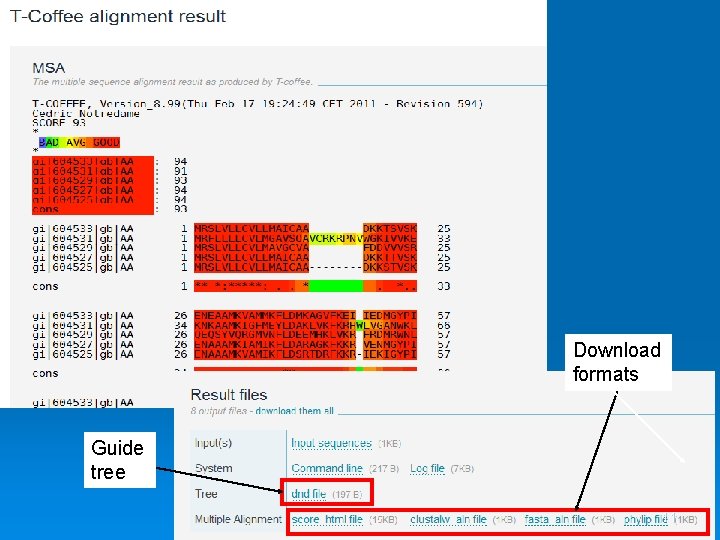
Download formats Guide tree 11

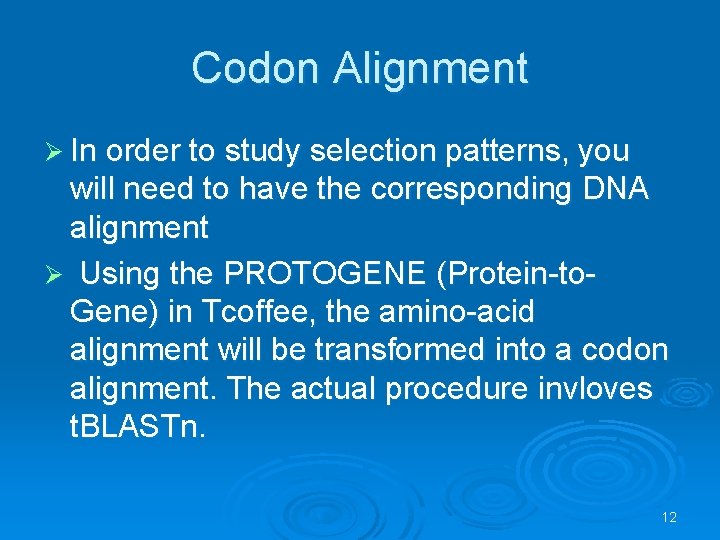
Codon Alignment Ø In order to study selection patterns, you will need to have the corresponding DNA alignment Ø Using the PROTOGENE (Protein-to. Gene) in Tcoffee, the amino-acid alignment will be transformed into a codon alignment. The actual procedure invloves t. BLASTn. 12

• PROTOGENE (in Tcoffee) is time consuming. Please submit your email address, and the results will be emailed to you. • PROTOGENE may return more that one DNA sequence for any given Protein sequence. For your homework assignment, please choose one sequence for each species. 13

(Result) Codon alignment >gi|604533|gb|AAC 37231. 1|_G_L 36554 _S_ AAC 37231 _DESC_ fertilization protein MATCHES_ON Haliotis assimilis fertilization protein m. RNA, complete cds ATGAGGTCTTTGGTGCTTCTCTGTGTTTTGCTGATGGCAATATGTGCGGCGGAC------------AAAAAAACCTCGGTCTCGAAGGAAAATGAAGCCGCAATGAAG GTAGCGATGATGAAGTTTTTGGATATGAAGGCGGGTGTATTCAAAGAAATC---ATTGAG GATATGGGATATCCAATAACCCCTCCGCAATGGACAACTCTACTGTACTACAACAGAGAG AGATTGAATTTTGCCGTTCCTTGCATTGTCCAAAAAGATTATATTGCTGGGA GGTAACAAATTAAATAAGGCGAACTTCGCTAGGATGGGTCGAATCCTTGGCTGGAAAAGC CAGTGGGCTGTGAGACAGAGGCAATGGGGGATGGTCAGA-----GTGTCGAGGCGC CATACAAGTACTGCAATAGCTAAAAGGATCGTCGCCATGAAAGTTGCTGACCTACCCTGT AAC---------TAG >gi|604531|gb|AAC 37233. 1|_G_L 36590 _S_ AAC 37233 _DESC_ fertilization protein MATCHES_ON Haliotis corrugata fertilization protein m. RNA, complete cds ATGAGGTTTTTGCTGCTTCTCTGTGTTTTGATGGGGGCAGTATCTCAGGCAGTATGCAGA AAAAGACCTAATGTCTGGGGGAAAATCGTGGTCAAGGAGAAAAATAAAGCCGCAATGAAG ATAGGGTTTATGGAATATTTGGATGCAAAGTTGGTAAAGTTTAAAAGGCACTGGCTTGTT GGAGCCAATTGGAAACTTCAAAAATTTGAAACGGATGAAATGAGATACCTCGCCATAAAG AGACTGATAAAAGTTTGCCATGGATACACTATTTGGTCCCAACGACTAATAATGTTAAAA TATCGACCATTGAATGAGAAATACTTCAAAAAGGTGGGTCGATACCTTGCCTGGCGAAAC TACCTCATAGTTTTTCGGATGTGGATCGGCGTTTTG------AAGAAAAATCTTAAAAGA TCGGAAATAACGAAACCCATGCAAAAACTCCTCGACACAAAGGATGGTGAGTTGCCCTGC CCTGTTAGAAAGATACATGGATAA >gi|604529|gb|AAC 37232. 1|_G_L 36589 _S_ AAC 37232 _DESC_ fertilization protein MATCHES_ON Haliotis fulgens fertilization protein m. RNA, complete cds ATGAGGTCTTTGGTGCTTCTCTGTGTTTTGATGGCGGTAGGATGTGTGGCGTTT------------GATGATGTGGTGGTCTCAAGGCAAGAGCAATCTTATGTGCAG AGAGGGATGGTCAACTTTTTGGATGAAGAAATGCATAAACTGGTTAAACGG---TTTAGA GATATGCGATGGAATTTAGGGCCAGGCTTTGTATTCCTTCTAAAAAAAGTCAACAGAGAG AGAATGATGCGCTACTGCATGGATTACGCCAGATATTCCAAAAAGATTTTACAGCTAAAA CATCTTCCAGTAAATAAGAAGACCCTCACTAAAATGGGTAGATTCGTTGGATATCGAAAC TATGGGGTCATCAGGGAGTTGTACGCCGACGTATTCAGAGACGTTCAAGGATTTAGGGGG CCTAAAATGACTGCAGCCATGAGGAAGTACAGCAGCAAGGATCCTGGTACATTTCCTTGC AAGAACGAGAAACGCCGCGGATGA >gi|604527|gb|AAC 37230. 1|_G_L 36553 _S_ AAC 37230 _DESC_ fertilization protein MATCHES_ON Haliotis sorenseni fertilization protein m. RNA, complete cds ATGAGGTCTTTGGTGCTTCTCTGTGTTTTGCTGATGGCAATATGTGCGGCGGAC------------AAAAAAACCACGGTCTCGAAGGAAAATGCAGCCGCAATGAAG ATAGCTATGATAAAGTTTTTGGATGCGAGGGCGGGTAAATTCAAAAAACGC---GTTGAG AATATGGGATATCCAATAACCCCTCCGCAATGGACAACTCTACTACAACAG AGATTGATGGAATGGTGCCATACCTACGTTGAATTTTCCAAAAAGATTATATTGATGGGA GGTAACAAATTAAATAAGAAGAACTTCACTAGGATGGGTCGAATCATTGGCTGGAAAAAC CAGTGGGTTTTGAAAAGGAGGCAATGGGAGATGGTCAGA-----GTGATGAGGCGC TATAAAAGTACTGCAATAGCTAAAAAGATCGTCGCCATGAAAGTTGCTGACCTACCCTGT AAC---------TAG >gi|604525|gb|AAC 37229. 1|_G_L 36552 _S_ AAC 37229 _DESC_ fertilization protein MATCHES_ON Haliotis rufescens fertilization protein m. RNA, complete cds ATGAGGTCTTTGGTGCTTCTCTGTGTTTTGCTGATGGCAATATGTGCGGCGGAC------------AAAAAATCCACGGTCTCGAAGGAAAATGCAGCCGCAATGAAG GTAGCGATGATAAAGTTTTTGGATTCGAGGACGGATAGATTCAAAAAACGC---ATTGAG AAGATTGGATATCCAATAACCCCTCCGCAATATACAACTCTACTACAACAGAGAG AGATTGATGGATTGGTGCCATAACTACGTTGAAGTATCCAAAAAGATTATATTGTTGGGA GGTAACAAATTAAATAAGAAGAACTTCGCTAGGATGGGTCGAATCATTGGCTGGAAAAAC CAGTGGATTTTGAAAAGGAGGCAATGGCACATGGTCAGA-----GTGATGAGGCGC TATAAAGCTTCTGCAATAGCTAAAAAGATCGTCGCCATGAAAGTTGCTGACCTACCCTGT AAC---------TAG 14

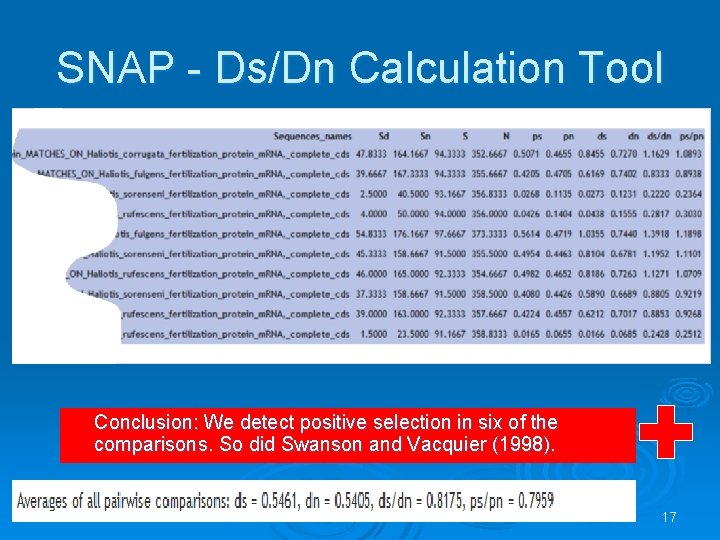
SNAP - Ds/Dn Calculation Tool http: //hcv. lanl. gov/content/sequence/SNAP. html Calculates synonymous and nonsynonymous substitution rates based on codon alignments according to Nei and Gojobori (1986) method. 15

Input codon alignment Select output statistics 16

SNAP - Ds/Dn Calculation Tool Conclusion: We detect positive selection in six of the comparisons. So did Swanson and Vacquier (1998). 17

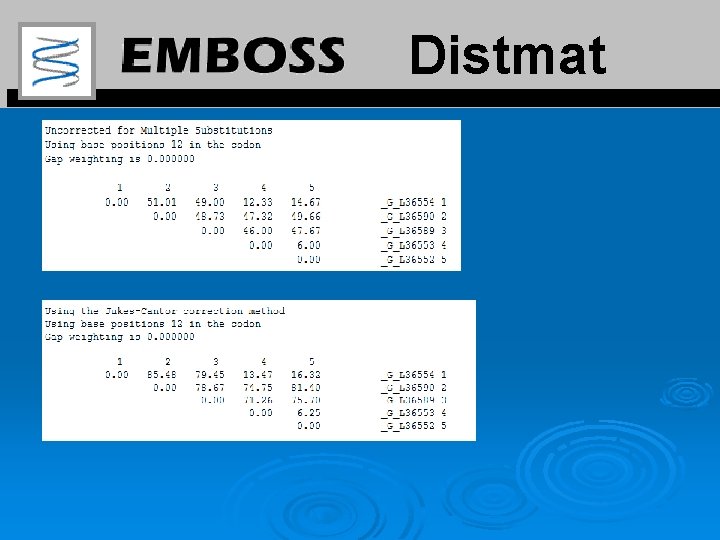
Distmat http: //emboss. bioinformatics. nl/cgi-bin/emboss/distmat Distmat calculates the evolutionary distances between every pair of sequences in a multiple alignment. The distances are expressed in terms of the number per 100 nucleotides or number of replacements per 100 amino acids 18

Distmat Ø Feed the DNA alignment of 18 -k. Da protein into distmat. Ø Calculate separately the distances between the sequences for codon positions 1 and 2, and for codon position 3. Ø Are the results in agreement with those from the dn/ds analysis? 19

Distmat

Distmat

Anchored multiple-sequence alignment with DIALIGN http: //dialign. gobics. de/anchor/submission. php User manual: http: //dialign. gobics. de/anchor/manual 22

Align the following sequences (use the file dalign_sequences. txt): >seq 1 WKKNADAPKRAMTSFMKAAY >seq 2 WNLDTNSPEEKQAYIQLAKDDRIRYD >seq 3 WRMDSNQKNPDSNNPKAAYNKGDANAPK 23

Results Ø DIALIGN makes alignments from fragments 24

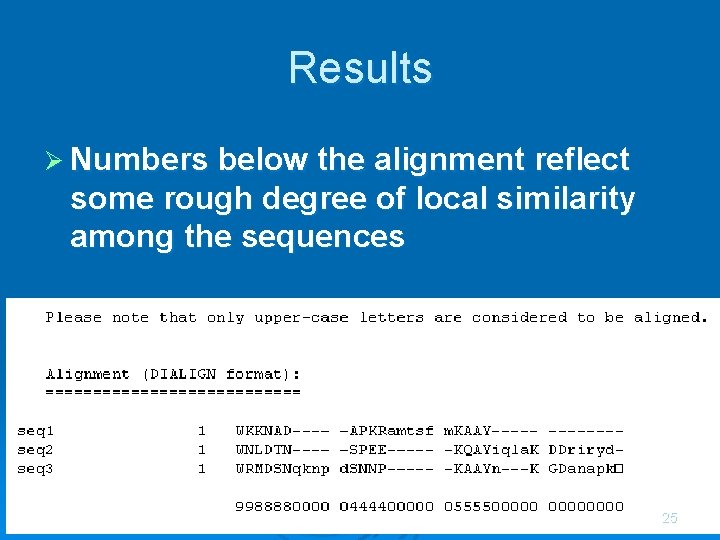
Results Ø Numbers below the alignment reflect some rough degree of local similarity among the sequences 25

Anchored alignment Ø Now, let us assume that the user has some expert knowledge concerning a certain domain that is present in all the input sequences Ø The domains marked in red in the three sequences are thought to be homologous to one another >seq 1 WKKNADAPKRAMTSFMKAAY >seq 2 WNLDTNSPEEKQAYIQLAKDDRIRYD >seq 3 WRMDSNQKNPDSNNPKAAYNKGDANAPK 26

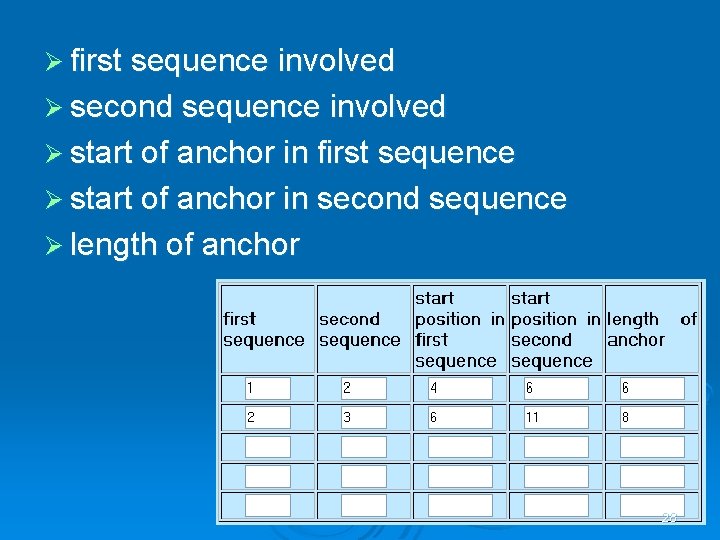
Ø Therefore, the user wants to define this domain as anchor and align the rest of the sequences automatically. Ø To specify a set of anchor points, each anchor point corresponds to a equal-length segment pair involving two of the input sequences should be defined 27

Ø first sequence involved Ø second sequence involved Ø start of anchor in first sequence Ø start of anchor in second sequence Ø length of anchor 28

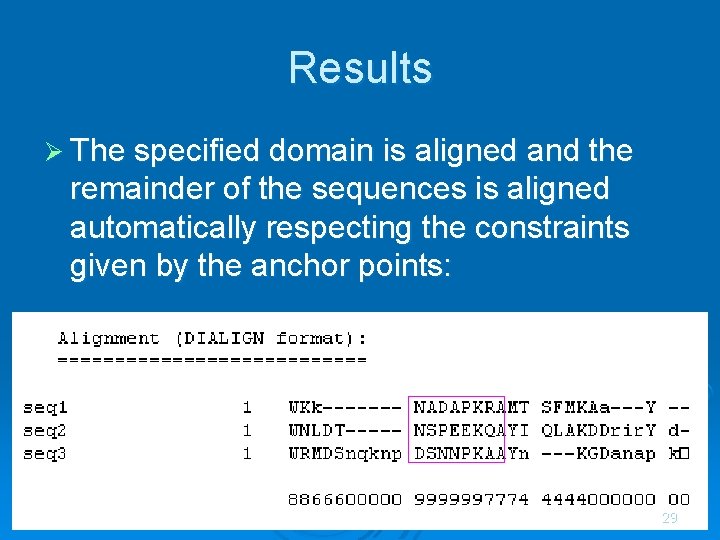
Results Ø The specified domain is aligned and the remainder of the sequences is aligned automatically respecting the constraints given by the anchor points: 29

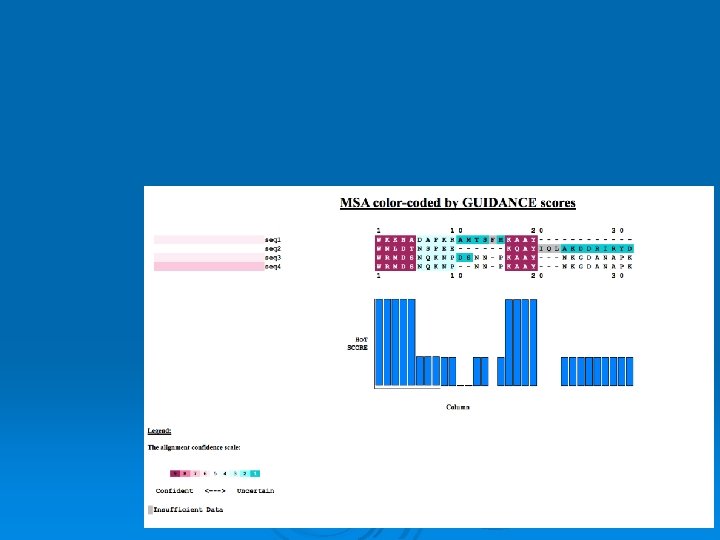
Guidance/Ho. T

>seq 1 WKKNADAPKRAMTSFMKAAY >seq 2 WNLDTNSPEEKQAYIQLAKDDRIRYD >seq 3 WRMDSNQKNPDSNNPKAAYNKGDANAPK >seq 4 WRMDSNQKNPNNPKAAYNKGDANAPK
