Multiple Sequence Alignment An alignment of heads Sequence




- Slides: 12

Multiple Sequence Alignment

An alignment of heads

Sequence Alignment • A way of arranging the primary sequences of DNA, RNA and amino acid to identify the regions of similarity that may be a consequence of functional, structural or evolutionary relationship between the sequences.

Goals • To establish an hypothesis of positional homology between bases/amino acids. • To generate a concise, information-rich summary of sequence data. • Sometimes used to illustrate the dissimilarity between a group of sequences. • Alignments can be treated as models that can be used to test hypotheses.

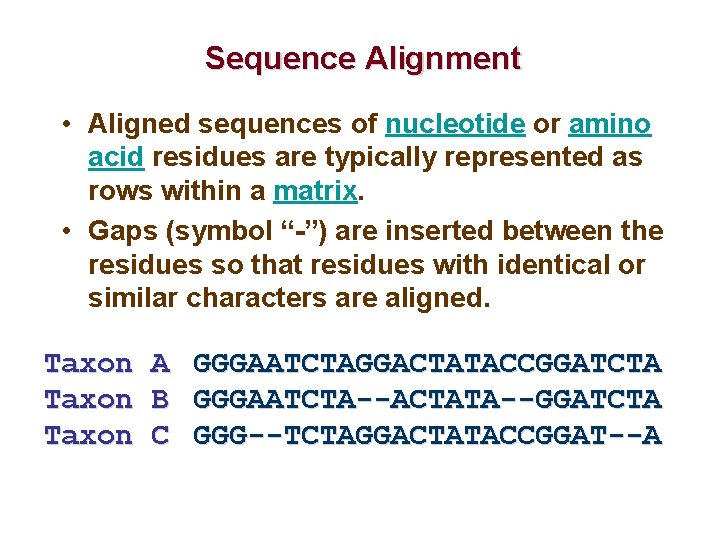
Sequence Alignment • Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. • Gaps (symbol “-”) are inserted between the residues so that residues with identical or similar characters are aligned. Taxon A GGGAATCTAGGACTATACCGGATCTA Taxon B GGGAATCTA--ACTATA--GGATCTA Taxon C GGG--TCTAGGACTATACCGGAT--A

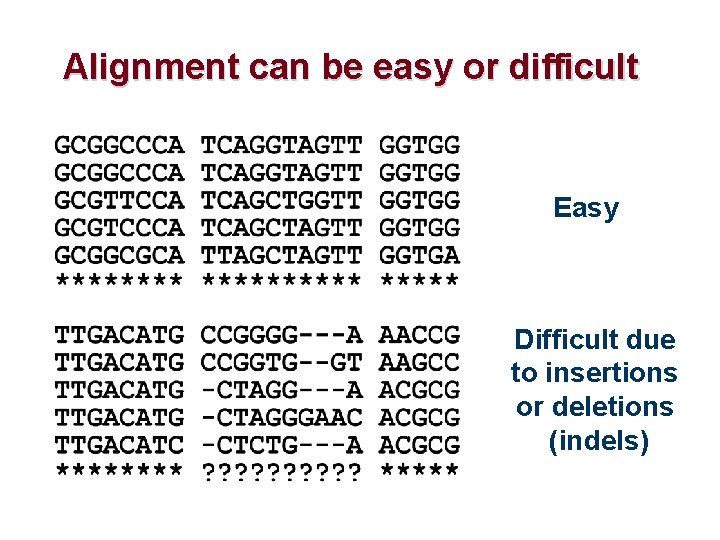
Alignment can be easy or difficult Easy Difficult due to insertions or deletions (indels)


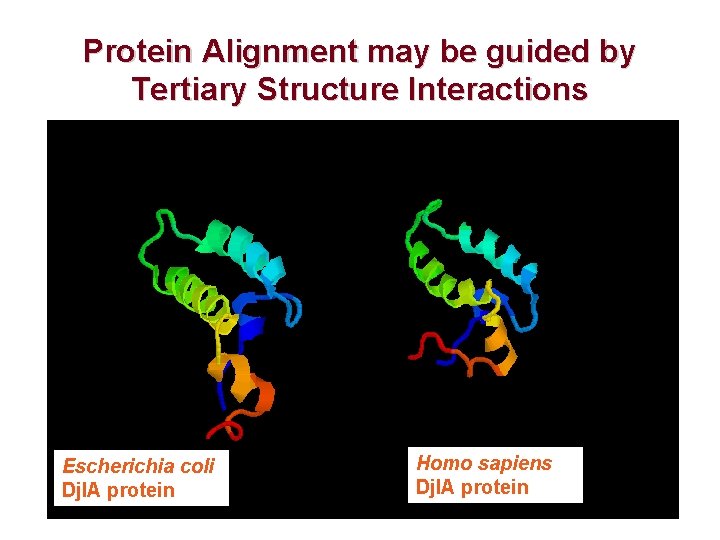
Protein Alignment may be guided by Tertiary Structure Interactions Escherichia coli Djl. A protein Homo sapiens Djl. A protein

Multiple Sequence Alignment. Approaches 3 main approaches of alignment: - Manual - Automatic - Combined

Manual Alignment Might be carried out because: - Alignment is easy. - There is some extraneous information (structural). - Automated alignment methods have encountered the local minimum problem. - An automated alignment method can be “improved”.

Automatic Alignment: Progressive Approach • Devised by Feng and Doolittle in 1987. • Essentially a heuristic method and as such is not guaranteed to find the ‘optimal’ alignment. • Requires n-1+n-2+n-3. . . n-n+1 pairwise alignments as a starting point. • Most successful implementation is CLUSTAL.

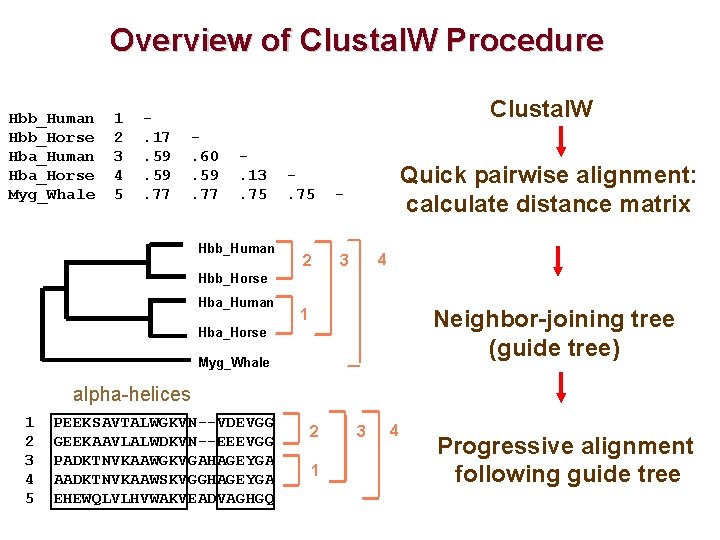
Overview of Clustal. W Procedure Hbb_Human Hbb_Horse Hba_Human Hba_Horse Myg_Whale 1 2 3 4 5 . 17. 59. 77 Clustal. W. 60. 59. 77 . 13. 75 Hbb_Human . 75 - 2 3 Quick pairwise alignment: calculate distance matrix 4 Hbb_Horse Hba_Human Neighbor-joining tree (guide tree) 1 Hba_Horse Myg_Whale alpha-helices 1 2 3 4 5 PEEKSAVTALWGKVN--VDEVGG GEEKAAVLALWDKVN--EEEVGG PADKTNVKAAWGKVGAHAGEYGA AADKTNVKAAWSKVGGHAGEYGA EHEWQLVLHVWAKVEADVAGHGQ 2 1 3 4 Progressive alignment following guide tree