Mechanisms of acute toxicity and using Tox Cast



















- Slides: 72

Mechanisms of acute toxicity and using Tox. Cast data to predict acute toxicity Dan Wilson, Ph. D, DABT Science Leader – Cheminformatics The Dow Chemical Company ddwilson@dow. com (989)636 -0712 EPA’s Computational Toxicology Communities of Practice Oct 22, 2015

Outline • • • How did we come to focus on this research? Acute toxicity regulatory landscape Potential mechanisms of high acute toxicity Our research Future opportunities

Acknowledgements …Back to the Future II ? Were AOPs 1 st mentioned? ?

Acknowledgements • Dow Chemical • EPA/NTP/NIEHS – Tyler Auernhammer – Mike Bartels – Barun Bhhatarai – Greg Bond • – Ed Carney – Shubhra • Chaudhuri – Bhaskar Gollapudi – Amanda Parks • – Paul Price – Pam Spencer – – Linda Birnbaum Warren Casey Richard Judson Bob Kavlock NCATS – Menghang Xia Simulations Plus – John Di. Bella PETA/PCRM – Amy Clippinger – Kristie Sullivan

Acknowledgements • Dow Chemical • EPA/NTP/NIEHS – Tyler Auernhammer – Mike Bartels – Barun Bhhatarai – Greg Bond • – Ed Carney – Shubhra • Chaudhuri – Bhaskar Gollapudi – Amanda Parks • – Paul Price – Pam Spencer – – Linda Birnbaum Warren Casey Richard Judson Bob Kavlock NCATS – Menghang Xia Simulations Plus – John Di. Bella PETA/PCRM – Amy Clippinger – Kristie Sullivan

Acknowledgements • Dow Chemical • EPA/NTP/NIEHS – Tyler Auernhammer – Mike Bartels – Barun Bhhatarai – Greg Bond • – Ed Carney – Shubhra • Chaudhuri – Bhaskar Gollapudi – Amanda Parks • – Paul Price – Pam Spencer – – Linda Birnbaum Warren Casey Richard Judson Bob Kavlock NCATS – Menghang Xia Simulations Plus – John Di. Bella PETA/PCRM – Amy Clippinger – Kristie Sullivan

Toxicity Study Types • Acute Tox – – – Oral Toxicity Dermal Toxicity Inhalation Toxicity Dermal Corrosion Eye Corrosion Skin Sensitization • Repeated Dose Tox – Rat, mouse, dog – 28 -day, 90 -day, chronic – Oral, dermal, inhalation • Genetic Toxicity • Cancer Bioassay – Lifetime study of rat/mouse • Developmental teratology – Potential for birth defects • Reproductive Tox – Effects on M/F fertility – Effects on the offspring – Multigenerational effects • Neuro Tox • Immunology • ADME (Absorption, Distribution, Metabolism, Excretion)

Background: Focus on acute tox • 2007: NAS report Toxicity Testing in 21 st Century • 2013: Linda Birnbaum publishes EHP editorial redirecting ICCVAM to focus on HTS and computational approaches • 2013: ICCVAM establishes acute toxicity as 1 of 3 strategic near-term priority areas for enhanced, immediate resource investment with expectation of short-term success • Acute toxicity should be a simpler and easier endpoint

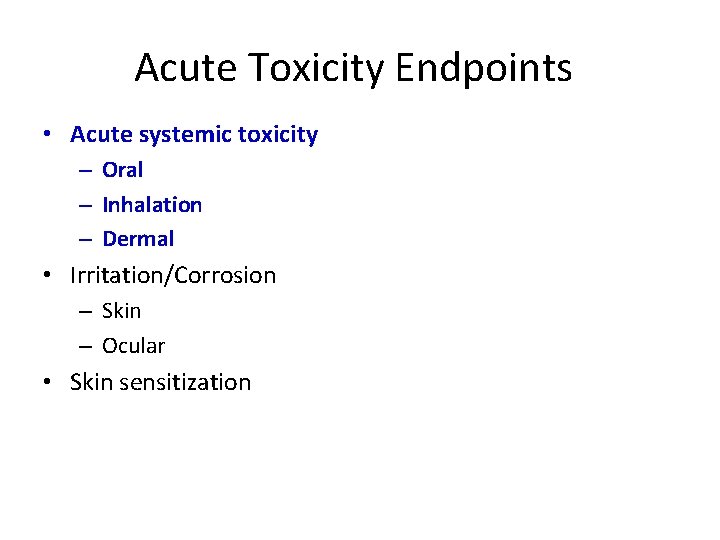
Acute Toxicity Endpoints • Acute systemic toxicity – Oral – Inhalation – Dermal • Irritation/Corrosion – Skin – Ocular • Skin sensitization

Where is acute toxicity testing required? USA • EPA - Pesticides • FDA - Medical devices EU-REACH • New registered substances >1 MT China • New registered substances >1 MT Korea • New registered substances >1 MT Taiwan • New registered substances >1 MT

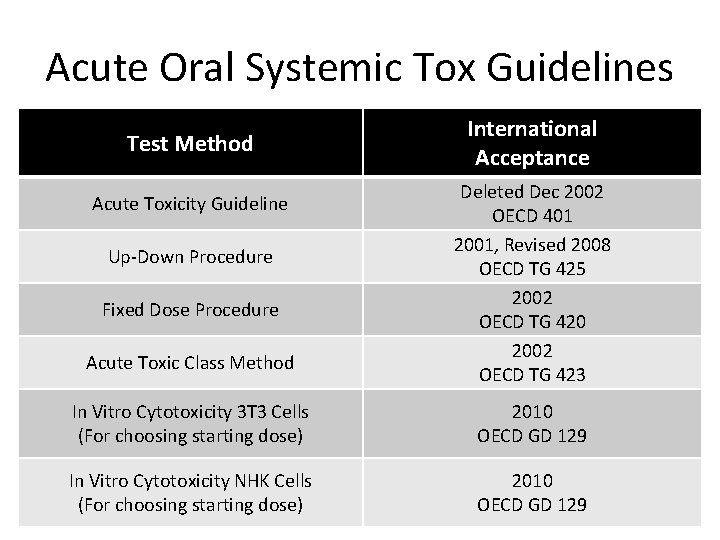
Acute Oral Systemic Tox Guidelines Test Method Acute Toxicity Guideline Up-Down Procedure Fixed Dose Procedure Acute Toxic Class Method International Acceptance Deleted Dec 2002 OECD 401 2001, Revised 2008 OECD TG 425 2002 OECD TG 420 2002 OECD TG 423 In Vitro Cytotoxicity 3 T 3 Cells (For choosing starting dose) 2010 OECD GD 129 In Vitro Cytotoxicity NHK Cells (For choosing starting dose) 2010 OECD GD 129

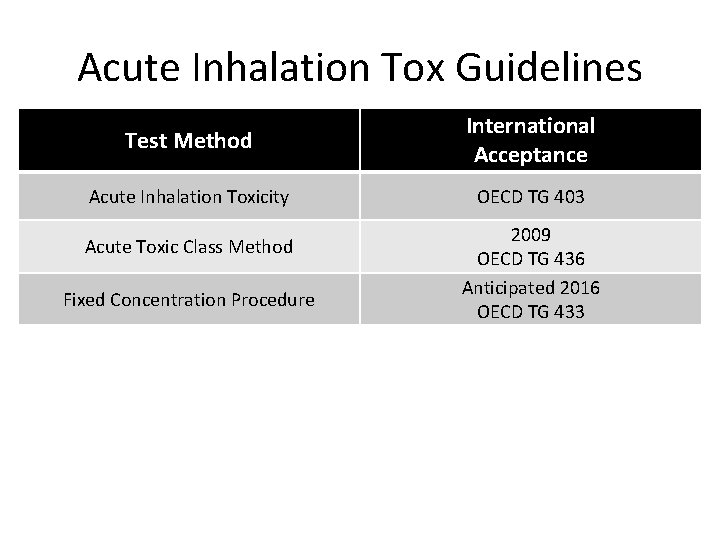
Acute Inhalation Tox Guidelines Test Method International Acceptance Acute Inhalation Toxicity OECD TG 403 Acute Toxic Class Method Fixed Concentration Procedure 2009 OECD TG 436 Anticipated 2016 OECD TG 433

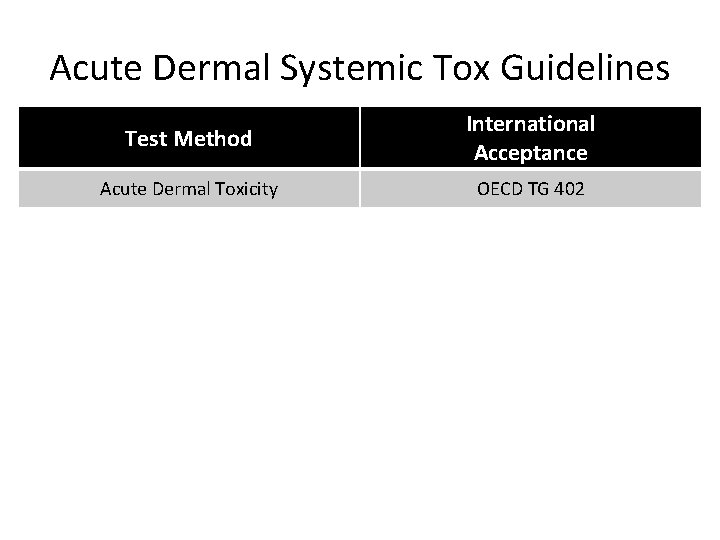
Acute Dermal Systemic Tox Guidelines Test Method International Acceptance Acute Dermal Toxicity OECD TG 402

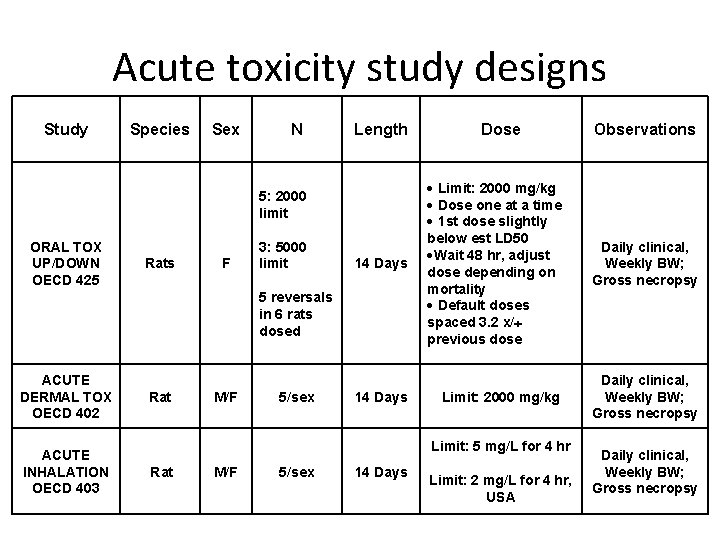
Acute toxicity study designs Study Species Sex N Length 5: 2000 limit ORAL TOX UP/DOWN OECD 425 Rats F 3: 5000 limit 14 Days 5 reversals in 6 rats dosed ACUTE DERMAL TOX OECD 402 ACUTE INHALATION OECD 403 Rat M/F 5/sex 14 Days Dose Limit: 2000 mg/kg Dose one at a time 1 st dose slightly below est LD 50 Wait 48 hr, adjust dose depending on mortality Default doses spaced 3. 2 x/ previous dose Daily clinical, Weekly BW; Gross necropsy Limit: 2000 mg/kg Daily clinical, Weekly BW; Gross necropsy Limit: 5 mg/L for 4 hr Rat M/F 5/sex 14 Days Observations Limit: 2 mg/L for 4 hr, USA Daily clinical, Weekly BW; Gross necropsy

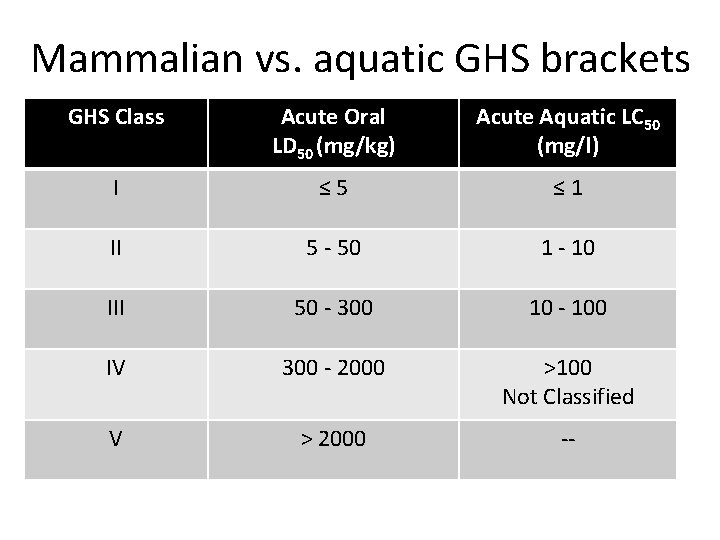
Mammalian vs. aquatic GHS brackets GHS Class Acute Oral LD 50 (mg/kg) Acute Aquatic LC 50 (mg/l) I ≤ 5 ≤ 1 II 5 - 50 1 - 10 III 50 - 300 10 - 100 IV 300 - 2000 >100 Not Classified V > 2000 --

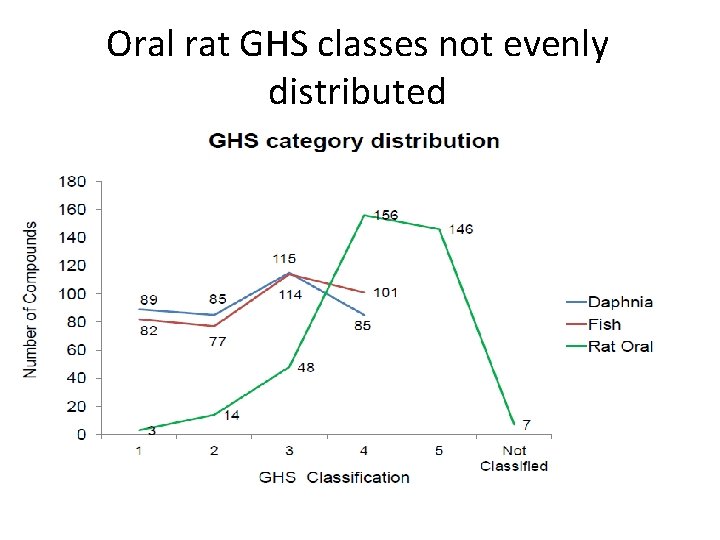
Oral rat GHS classes not evenly distributed

Regulations require to classify by LD/LC 50 • This can be done using guideline animal studies • Do in silico alternative methods offer a replacement? • Do in vitro alternative methods offer a replacement?

In silico approaches - QSAR

In Vitro Approaches • Basal cytotoxicity in 3 T 3 or NHK cells – Set starting dose for acute animal toxicity studies – Miss compounds acting via some mechanisms • Basal cytotoxicity in differentiated neuronal cell models – Axonopathy; Neurite outgrowth; Ach receptor signaling; Noradrenalin uptake; Cell membrane potential; Voltage-operated Ca++ channels • Small organisms – Zebra fish embryo – C. Elegans • High-throughput-screening data – Tox. Cast – Tox 21

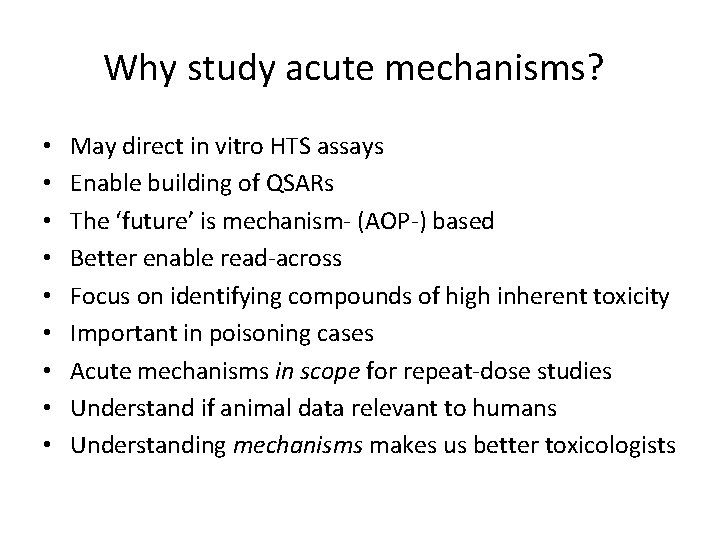
Why study acute mechanisms? • • • May direct in vitro HTS assays Enable building of QSARs The ‘future’ is mechanism- (AOP-) based Better enable read-across Focus on identifying compounds of high inherent toxicity Important in poisoning cases Acute mechanisms in scope for repeat-dose studies Understand if animal data relevant to humans Understanding mechanisms makes us better toxicologists

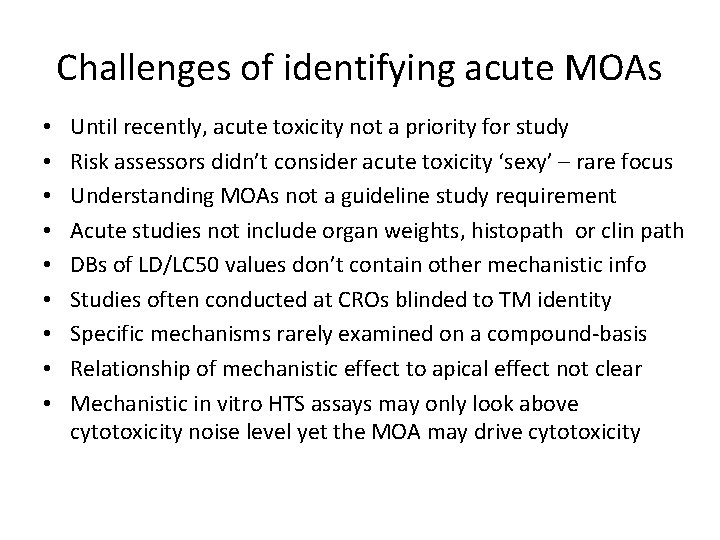
Challenges of identifying acute MOAs • • • Until recently, acute toxicity not a priority for study Risk assessors didn’t consider acute toxicity ‘sexy’ – rare focus Understanding MOAs not a guideline study requirement Acute studies not include organ weights, histopath or clin path DBs of LD/LC 50 values don’t contain other mechanistic info Studies often conducted at CROs blinded to TM identity Specific mechanisms rarely examined on a compound-basis Relationship of mechanistic effect to apical effect not clear Mechanistic in vitro HTS assays may only look above cytotoxicity noise level yet the MOA may drive cytotoxicity

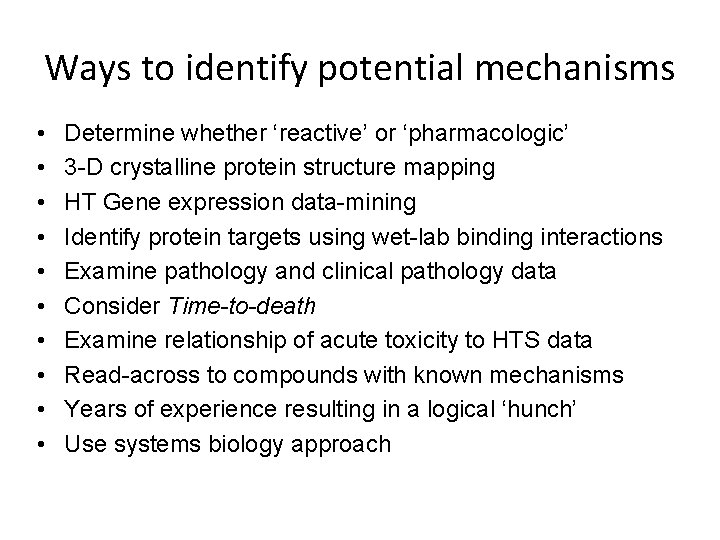
Ways to identify potential mechanisms • • • Determine whether ‘reactive’ or ‘pharmacologic’ 3 -D crystalline protein structure mapping HT Gene expression data-mining Identify protein targets using wet-lab binding interactions Examine pathology and clinical pathology data Consider Time-to-death Examine relationship of acute toxicity to HTS data Read-across to compounds with known mechanisms Years of experience resulting in a logical ‘hunch’ Use systems biology approach

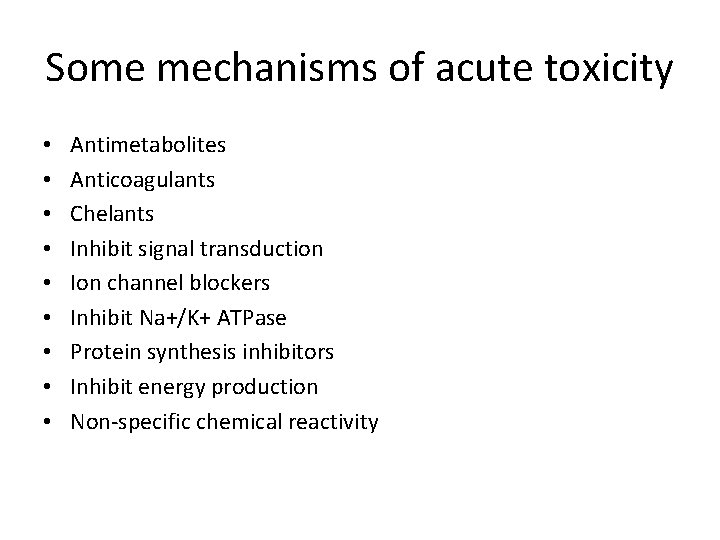
Some mechanisms of acute toxicity • • • Antimetabolites Anticoagulants Chelants Inhibit signal transduction Ion channel blockers Inhibit Na+/K+ ATPase Protein synthesis inhibitors Inhibit energy production Non-specific chemical reactivity

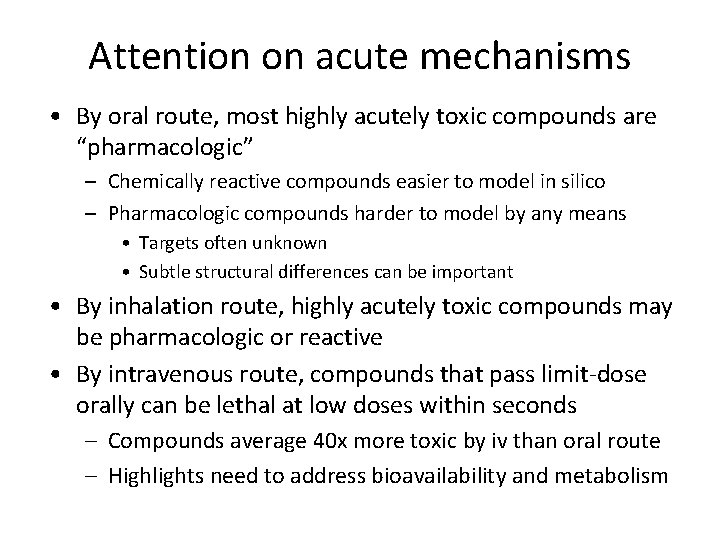
Attention on acute mechanisms • By oral route, most highly acutely toxic compounds are “pharmacologic” – Chemically reactive compounds easier to model in silico – Pharmacologic compounds harder to model by any means • Targets often unknown • Subtle structural differences can be important • By inhalation route, highly acutely toxic compounds may be pharmacologic or reactive • By intravenous route, compounds that pass limit-dose orally can be lethal at low doses within seconds – Compounds average 40 x more toxic by iv than oral route – Highlights need to address bioavailability and metabolism

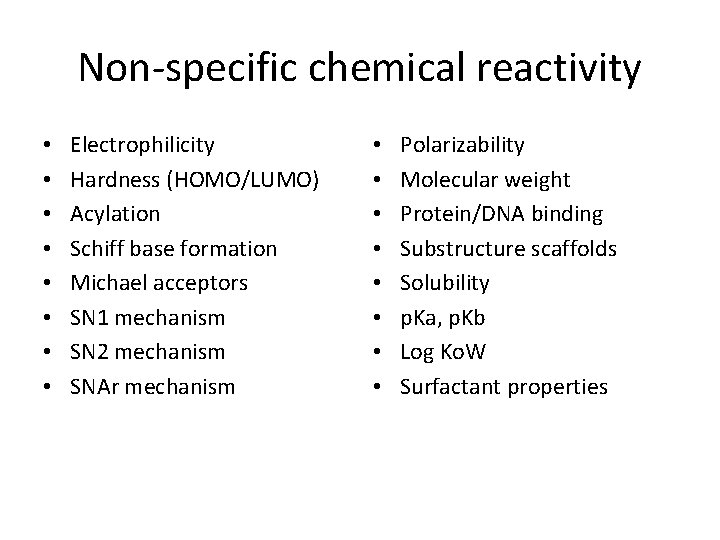
Non-specific chemical reactivity • • Electrophilicity Hardness (HOMO/LUMO) Acylation Schiff base formation Michael acceptors SN 1 mechanism SN 2 mechanism SNAr mechanism • • Polarizability Molecular weight Protein/DNA binding Substructure scaffolds Solubility p. Ka, p. Kb Log Ko. W Surfactant properties

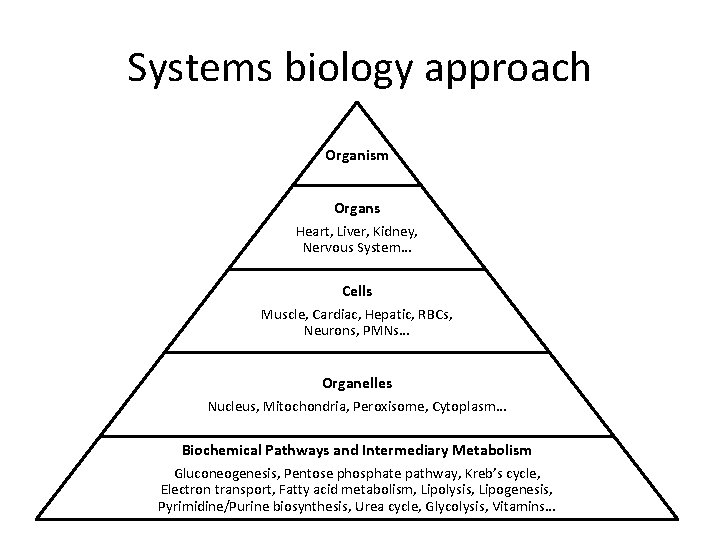
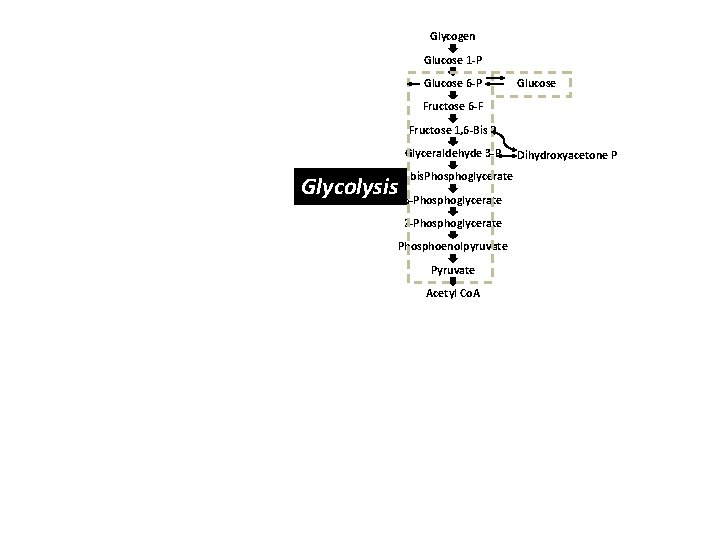
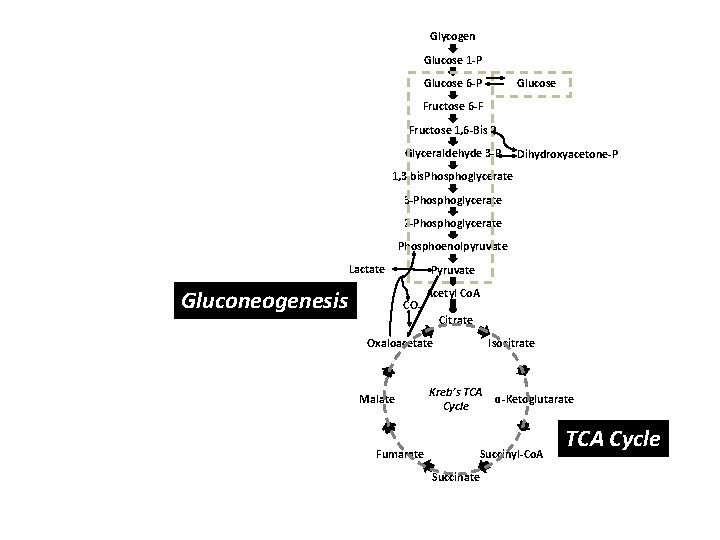
Systems biology approach Organism Organs Heart, Liver, Kidney, Nervous System… Cells Muscle, Cardiac, Hepatic, RBCs, Neurons, PMNs… Organelles Nucleus, Mitochondria, Peroxisome, Cytoplasm… Biochemical Pathways and Intermediary Metabolism Gluconeogenesis, Pentose phosphate pathway, Kreb’s cycle, Electron transport, Fatty acid metabolism, Lipolysis, Lipogenesis, Pyrimidine/Purine biosynthesis, Urea cycle, Glycolysis, Vitamins…

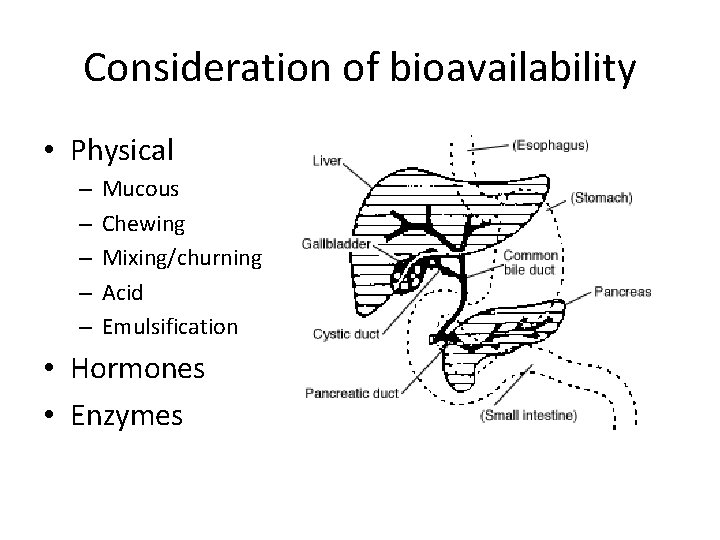
Consideration of bioavailability • Physical – – – Mucous Chewing Mixing/churning Acid Emulsification • Hormones • Enzymes

Protein Digestion • Stomach – HCl denatures – Pepsinogen → pepsin • Small Intestine – Hormones • Cholecystokinin • Secretin – Pancreatic enzymes • Trypsin, peptidases, elastase • Amino acids ↑ insulin, ↓ glucagon • No storage form for protein – amino acids → protein; carbons → carbohydrate/lipid; amino “N” as urea

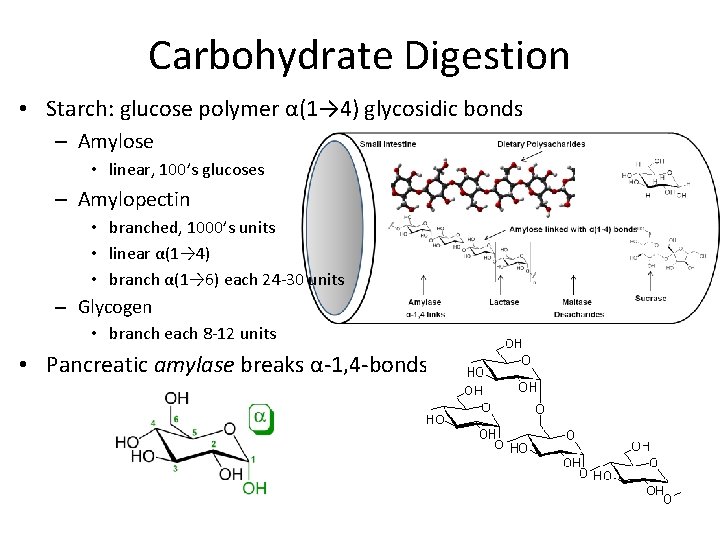
Carbohydrate Digestion • Starch: glucose polymer α(1→ 4) glycosidic bonds – Amylose • linear, 100’s glucoses – Amylopectin • branched, 1000’s units • linear α(1→ 4) • branch α(1→ 6) each 24 -30 units – Glycogen • branch each 8 -12 units • Pancreatic amylase breaks α-1, 4 -bonds

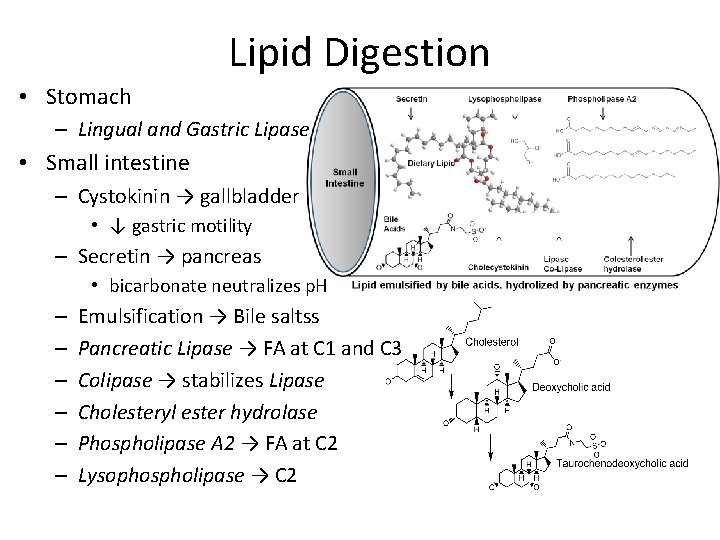
Lipid Digestion • Stomach – Lingual and Gastric Lipase • Small intestine – Cystokinin → gallbladder • ↓ gastric motility – Secretin → pancreas • bicarbonate neutralizes p. H – – – Emulsification → Bile saltss Pancreatic Lipase → FA at C 1 and C 3 Colipase → stabilizes Lipase Cholesteryl ester hydrolase Phospholipase A 2 → FA at C 2 Lysophospholipase → C 2

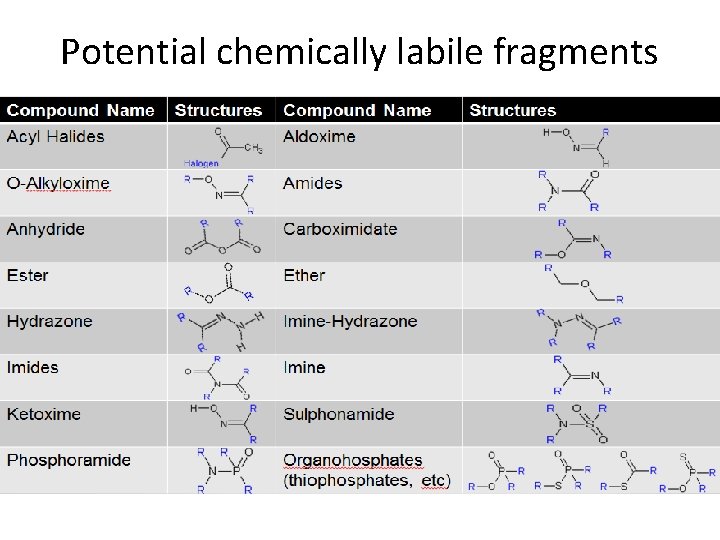
Potential chemically labile fragments

Glycogen Glucose 1 -P Glucose 6 -P Glucose Fructose 6 -F Fructose 1, 6 -Bis P Glyceraldehyde 3 -P 1, 3 bis. Phosphoglycerate Glycolysis 3 -Phosphoglycerate 2 -Phosphoglycerate Phosphoenolpyruvate Pyruvate Acetyl Co. A Dihydroxyacetone P

Glycogen Glucose 1 -P Glucose 6 -P Fructose 6 -F Fructose 1, 6 -Bis P Glyceraldehyde 3 -P Dihydroxyacetone-P 1, 3 bis. Phosphoglycerate 3 -Phosphoglycerate 2 -Phosphoglycerate Phosphoenolpyruvate Lactate Gluconeogenesis Pyruvate CO 2 Acetyl Co. A Citrate Oxaloacetate Malate Fumarate Isocitrate Kreb’s TCA Cycle ɑ-Ketoglutarate Succinyl-Co. A Succinate TCA Cycle

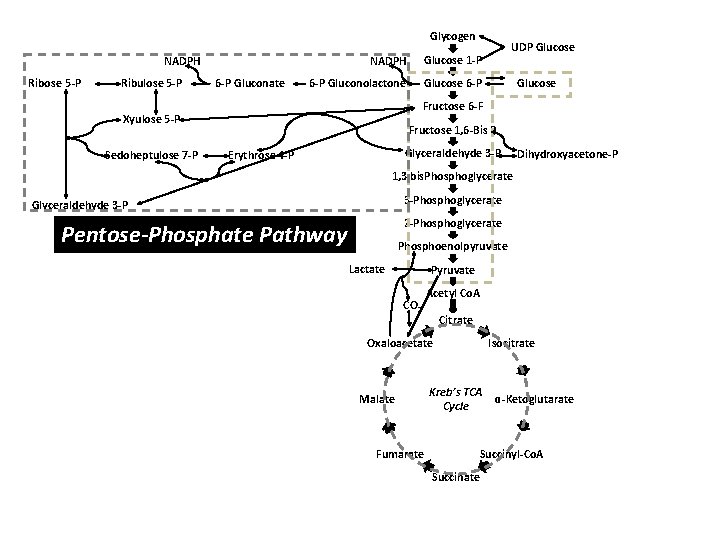
Glycogen NADPH Glucose 1 -P 6 -P Gluconolactone Glucose 6 -P NADPH Ribose 5 -P Ribulose 5 -P 6 -P Gluconate Glucose Fructose 6 -F Xyulose 5 -P Sedoheptulose 7 -P UDP Glucose Fructose 1, 6 -Bis P Glyceraldehyde 3 -P Erythrose 4 -P Dihydroxyacetone-P 1, 3 bis. Phosphoglycerate 3 -Phosphoglycerate Glyceraldehyde 3 -P 2 -Phosphoglycerate Pentose-Phosphate Pathway Phosphoenolpyruvate Lactate Pyruvate CO 2 Acetyl Co. A Citrate Oxaloacetate Malate Fumarate Isocitrate Kreb’s TCA Cycle ɑ-Ketoglutarate Succinyl-Co. A Succinate

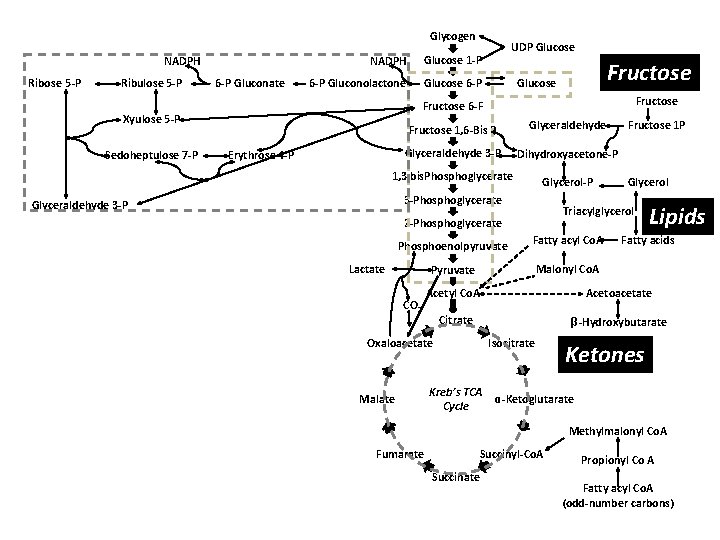
Glycogen NADPH Ribose 5 -P Ribulose 5 -P 6 -P Gluconate NADPH Glucose 1 -P 6 -P Gluconolactone Glucose 6 -P Fructose Glucose Fructose 6 -F Xyulose 5 -P Sedoheptulose 7 -P UDP Glucose Erythrose 4 -P Fructose 1, 6 -Bis P Glyceraldehyde 3 -P Dihydroxyacetone-P 1, 3 bis. Phosphoglycerate Glycerol-P 3 -Phosphoglycerate Glyceraldehyde 3 -P Lactate Fatty acyl Co. A Fatty acids Acetyl Co. A Acetoacetate Citrate β-Hydroxybutarate Oxaloacetate Malate Lipids Malonyl Co. A Pyruvate CO 2 Glycerol Triacylglycerol 2 -Phosphoglycerate Phosphoenolpyruvate Fructose 1 P Isocitrate Kreb’s TCA Cycle Ketones ɑ-Ketoglutarate Methylmalonyl Co. A Fumarate Succinyl-Co. A Succinate Propionyl Co A Fatty acyl Co. A (odd-number carbons)

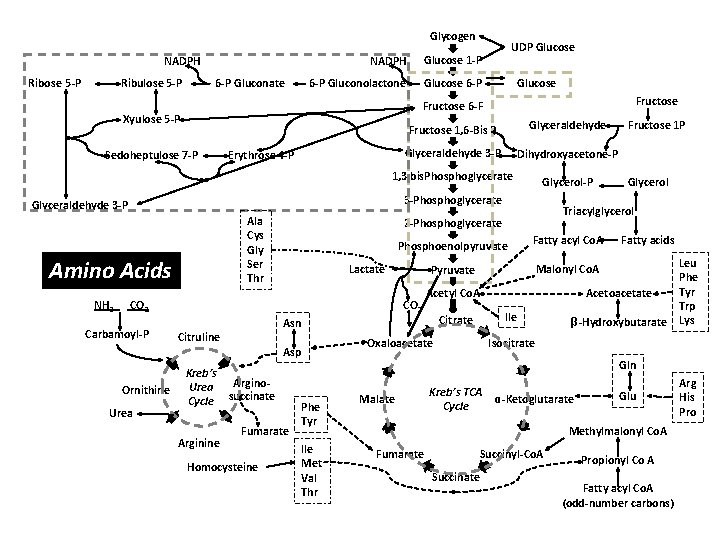
Glycogen NADPH Glucose 1 -P 6 -P Gluconolactone Glucose 6 -P NADPH Ribose 5 -P Ribulose 5 -P 6 -P Gluconate UDP Glucose Fructose 6 -F Xyulose 5 -P Sedoheptulose 7 -P Erythrose 4 -P Fructose 1, 6 -Bis P Glyceraldehyde 3 -P Dihydroxyacetone-P 1, 3 bis. Phosphoglycerate 3 -Phosphoglycerate Glyceraldehyde 3 -P Ala Cys Gly Ser Thr Amino Acids NH 3 Glycerol-P Phosphoenolpyruvate CO 2 Carbamoyl-P Asp Ornithine Urea Kreb’s Urea Cycle Arginine Arginosuccinate Fumarate Homocysteine Phe Tyr Ile Met Val Thr Fatty acids Malonyl Co. A Acetoacetate Ile Citrate Asn Citruline Fatty acyl Co. A Pyruvate CO 2 Glycerol Triacylglycerol 2 -Phosphoglycerate Lactate Fructose 1 P Oxaloacetate β-Hydroxybutarate Leu Phe Tyr Trp Lys Isocitrate Gln Malate Kreb’s TCA Cycle ɑ-Ketoglutarate Glu Methylmalonyl Co. A Fumarate Succinyl-Co. A Succinate Propionyl Co A Fatty acyl Co. A (odd-number carbons) Arg His Pro

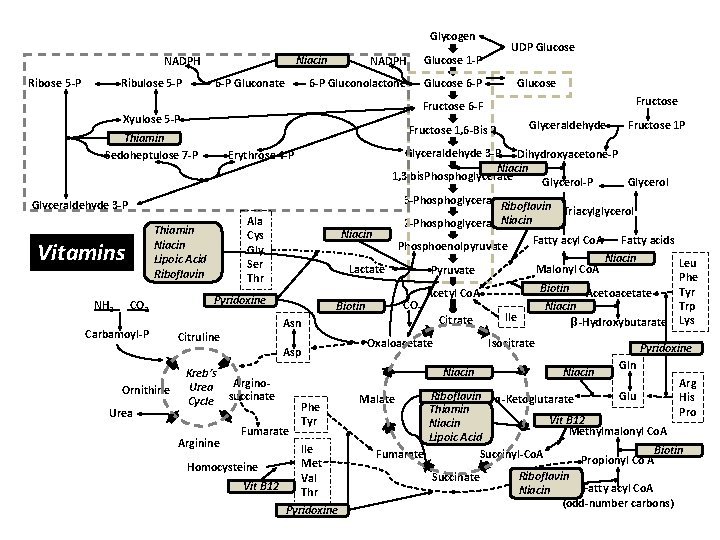
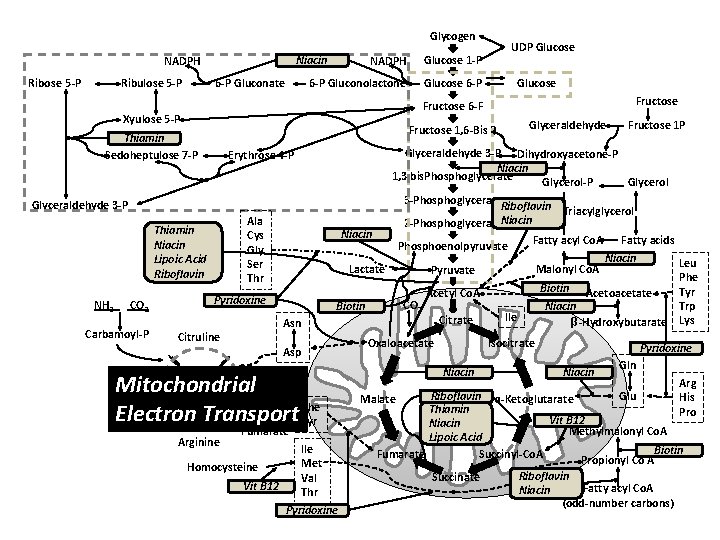
Glycogen Ribulose 5 -P 6 -P Gluconolactone Glucose 6 -P Niacin NADPH Ribose 5 -P NADPH Glucose 1 -P 6 -P Gluconate Ala Cys Gly Ser Thiamin Niacin Lipoic Acid Riboflavin Carbamoyl-P Niacin Pyridoxine Urea Arginine CO 2 Biotin Arginosuccinate Fumarate Ile Met Homocysteine Val Vit B 12 Thr Pyridoxine Fatty acids Niacin Leu Malonyl Co. A Phe Biotin Acetoacetate Tyr Trp Niacin β-Hydroxybutarate Lys Acetyl Co. A Citrate Oxaloacetate Ile Isocitrate Niacin Phe Tyr Fructose 1 P Fatty acyl Co. A Pyruvate Asn Citruline Kreb’s Urea Cycle Phosphoenolpyruvate Lactate Asp Ornithine Glyceraldehyde 3 -P Dihydroxyacetone-P Niacin 1, 3 bis. Phosphoglycerate Glycerol-P Glycerol 3 -Phosphoglycerate Riboflavin Triacylglycerol 2 -Phosphoglycerate. Niacin Erythrose 4 -P Glyceraldehyde 3 -P CO 2 Fructose 1, 6 -Bis P Thiamin Sedoheptulose 7 -P NH 3 Glucose Fructose 6 -F Xyulose 5 -P Vitamins UDP Glucose Pyridoxine Niacin Gln Arg His Pro Riboflavin ɑ-Ketoglutarate Glu Thiamin Vit B 12 Niacin Methylmalonyl Co. A Lipoic Acid Biotin Fumarate Succinyl-Co. A Propionyl Co A Malate Succinate Riboflavin Fatty acyl Co. A Niacin (odd-number carbons)

Glycogen Ribulose 5 -P 6 -P Gluconolactone Glucose 6 -P Niacin NADPH Ribose 5 -P NADPH Glucose 1 -P 6 -P Gluconate Glyceraldehyde Ala Cys Gly Ser Thiamin Niacin Lipoic Acid Riboflavin Niacin CO 2 Biotin Asp Kreb’s Argino. Urea Cycle succinate Mitochondrial Ornithine Urea Electron Transport Phe Tyr Fumarate Ile Met Homocysteine Val Vit B 12 Thr Pyridoxine Fatty acyl Co. A Fatty acids Niacin Leu Malonyl Co. A Phe Biotin Acetoacetate Tyr Trp Niacin β-Hydroxybutarate Lys Pyruvate Acetyl Co. A Citrate Asn Citruline Arginine Phosphoenolpyruvate Lactate Pyridoxine Fructose 1 P Glyceraldehyde 3 -P Dihydroxyacetone-P Niacin 1, 3 bis. Phosphoglycerate Glycerol-P Glycerol 3 -Phosphoglycerate Riboflavin Triacylglycerol 2 -Phosphoglycerate. Niacin Erythrose 4 -P Glyceraldehyde 3 -P Carbamoyl-P Fructose 1, 6 -Bis P Thiamin Sedoheptulose 7 -P CO 2 Glucose Fructose 6 -F Xyulose 5 -P NH 3 UDP Glucose Oxaloacetate Ile Isocitrate Niacin Pyridoxine Niacin Gln Arg His Pro Riboflavin ɑ-Ketoglutarate Glu Thiamin Vit B 12 Niacin Methylmalonyl Co. A Lipoic Acid Biotin Fumarate Succinyl-Co. A Propionyl Co A Malate Succinate Riboflavin Fatty acyl Co. A Niacin (odd-number carbons)

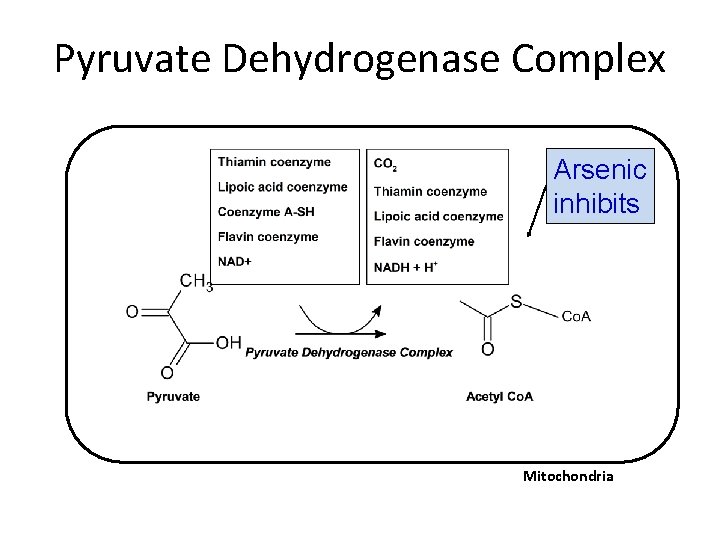
Pyruvate Dehydrogenase Complex Arsenic inhibits Mitochondria

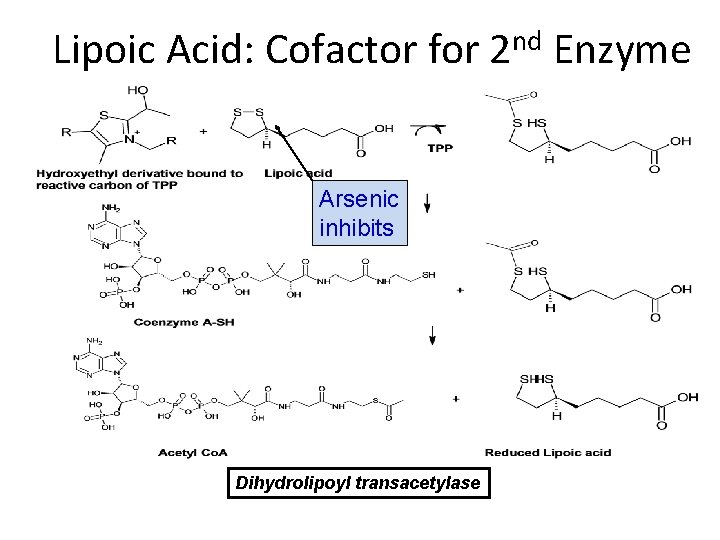
Lipoic Acid: Cofactor for 2 nd Enzyme Arsenic inhibits Dihydrolipoyl transacetylase

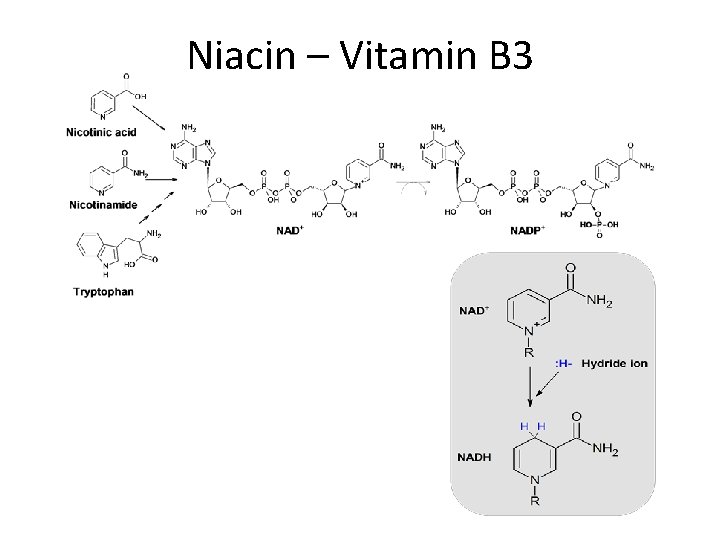
Niacin – Vitamin B 3

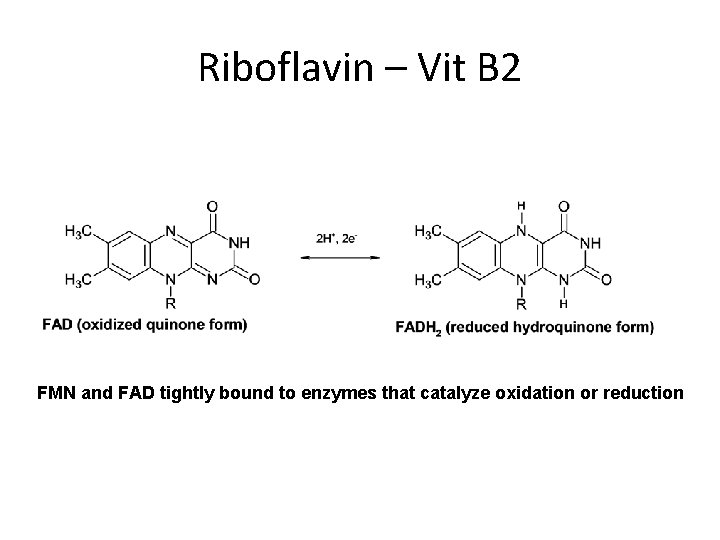
Riboflavin – Vit B 2 FMN and FAD tightly bound to enzymes that catalyze oxidation or reduction

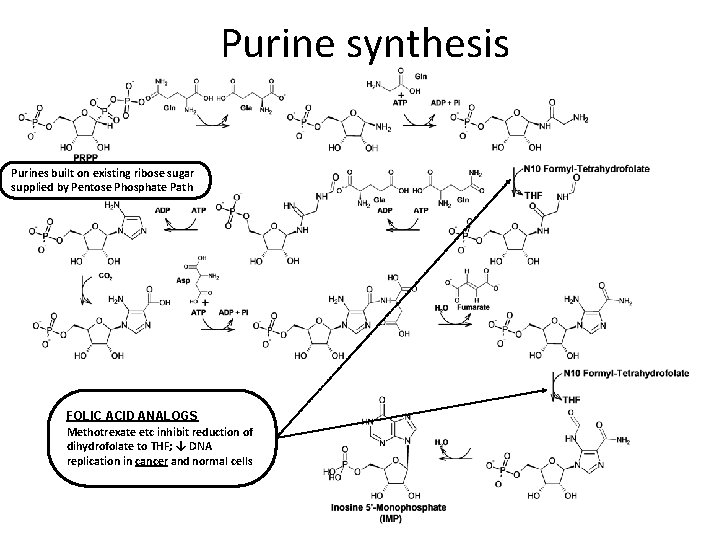
Purine synthesis Purines built on existing ribose sugar supplied by Pentose Phosphate Path FOLIC ACID ANALOGS Methotrexate etc inhibit reduction of dihydrofolate to THF; ↓ DNA replication in cancer and normal cells

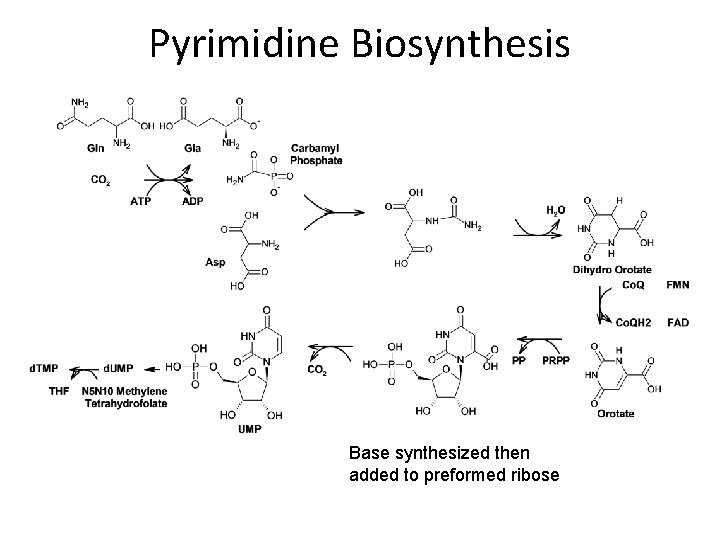
Pyrimidine Biosynthesis Base synthesized then added to preformed ribose

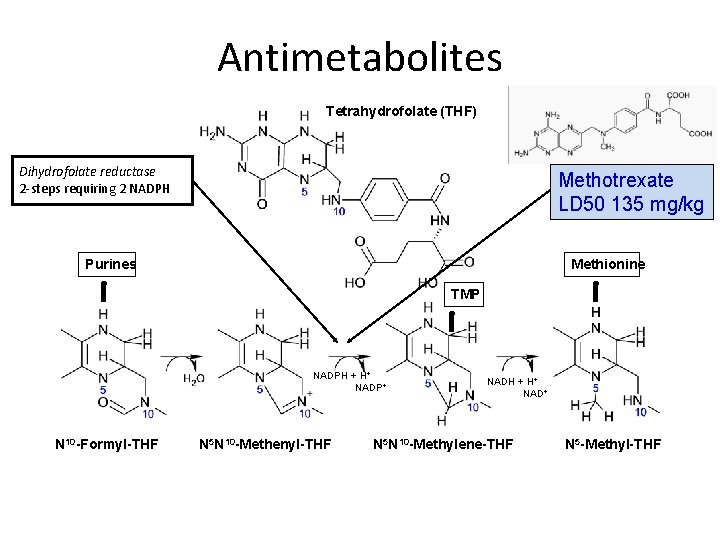
Antimetabolites Tetrahydrofolate (THF) Dihydrofolate reductase 2 -steps requiring 2 NADPH Methotrexate LD 50 135 mg/kg Purines Methionine TMP NADPH + H+ NADP+ N 10 -Formyl-THF N 5 N 10 -Methenyl-THF NADH + H+ NAD+ N 5 N 10 -Methylene-THF N 5 -Methyl-THF

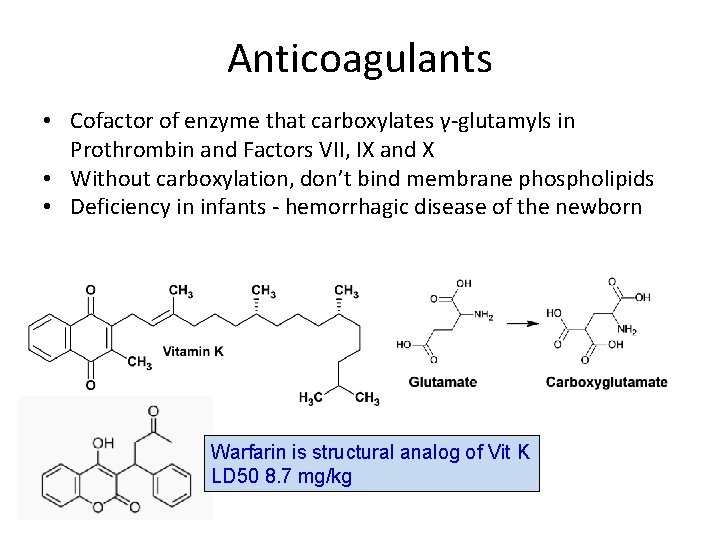
Anticoagulants • Cofactor of enzyme that carboxylates γ-glutamyls in Prothrombin and Factors VII, IX and X • Without carboxylation, don’t bind membrane phospholipids • Deficiency in infants - hemorrhagic disease of the newborn Warfarin is structural analog of Vit K LD 50 8. 7 mg/kg

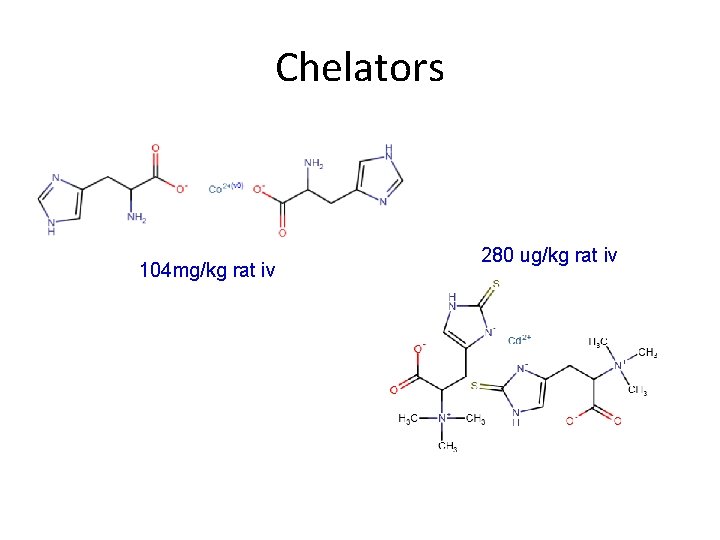
Chelators 104 mg/kg rat iv 280 ug/kg rat iv

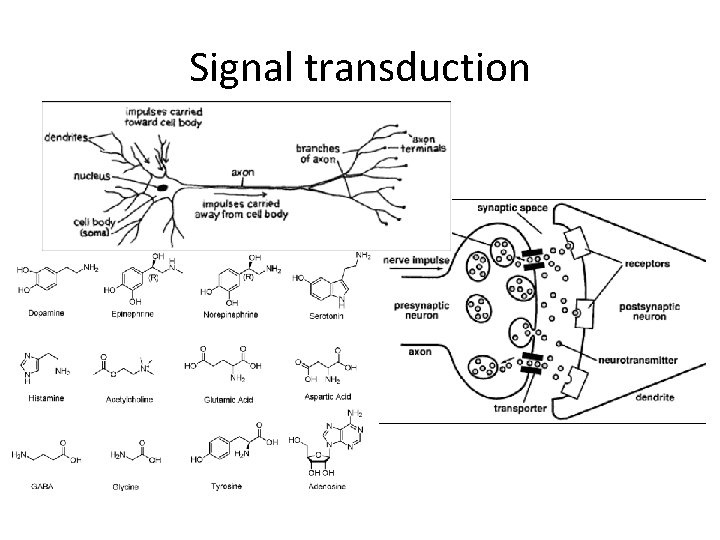
Signal transduction

Tetrodotoxin Inhibits voltage-gated sodium channels Oral LD 50 334 ug/kg

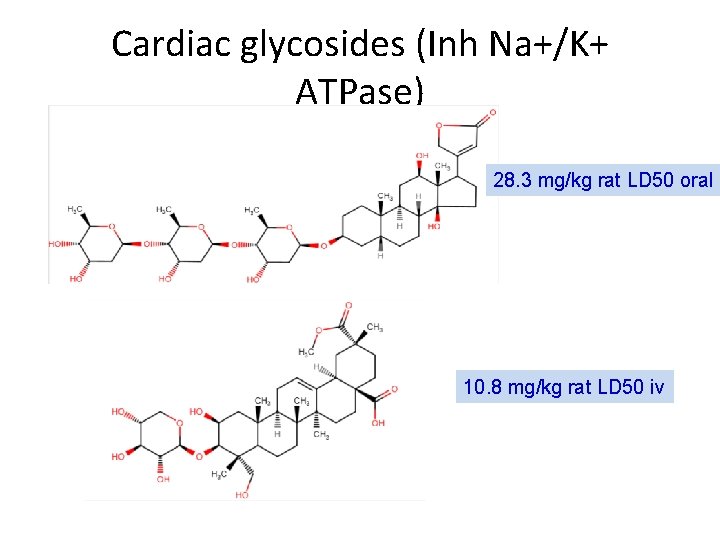
Cardiac glycosides (Inh Na+/K+ ATPase) 28. 3 mg/kg rat LD 50 oral 10. 8 mg/kg rat LD 50 iv

Michael acceptors Acrolein LD 50 26 mg/kg Methyl-methacrylate LD 50 7872 mg/kg

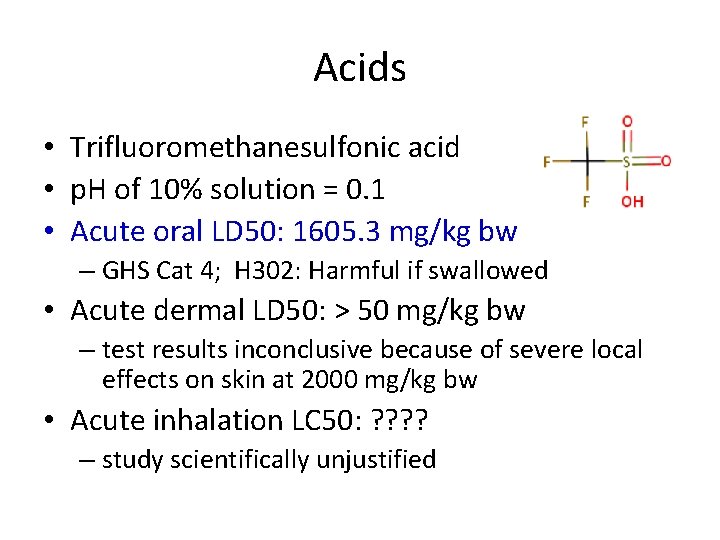
Acids • Trifluoromethanesulfonic acid • p. H of 10% solution = 0. 1 • Acute oral LD 50: 1605. 3 mg/kg bw – GHS Cat 4; H 302: Harmful if swallowed • Acute dermal LD 50: > 50 mg/kg bw – test results inconclusive because of severe local effects on skin at 2000 mg/kg bw • Acute inhalation LC 50: ? ? – study scientifically unjustified

Our Acute Toxicity Research • Catalogue and assess alternative models – In Vivo – In Vitro – In Silico • Compile toxicity databases – – Acute rat oral, intravenous, inhalation Acute daphnia, fish In vitro high-throughput screening data Eye and skin irritation/corrosion • Derive plausible AOPs/mechanisms • Address bioavailability and metabolism • Systems Biology approach to read-across

Alternative approaches to acute mammalian toxicity. Use of Tox. Cast™ mitochondrial inhibition data. PROOF-OF-CONCEPT

Current Hypothesis • Mechanisms of acute toxicity likely conserved across invertebrate, aquatic and mammalian species • Suggests dose-response concordance would be high and in vitro mechanistic data could predict responses in multiple species under conditions of similar bioavailability

Fish acute toxicity vs. Tox. Cast HTS

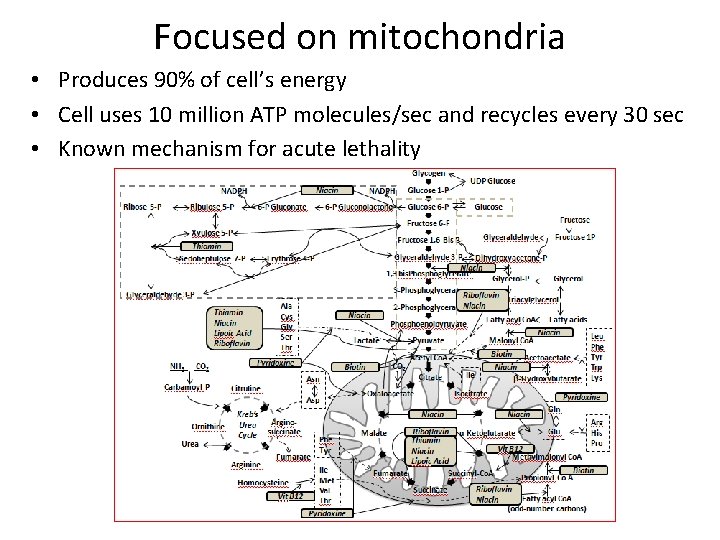
Focused on mitochondria • Produces 90% of cell’s energy • Cell uses 10 million ATP molecules/sec and recycles every 30 sec • Known mechanism for acute lethality

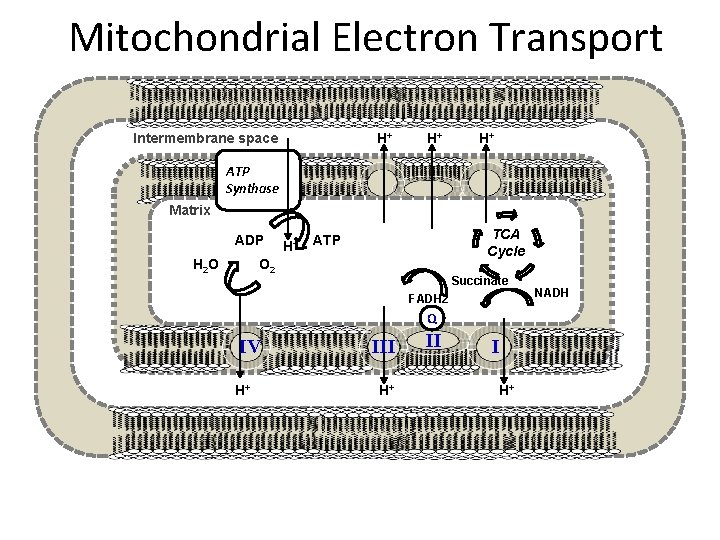
Mitochondrial Electron Transport Intermembrane space H+ H+ H+ ATP Synthase Matrix III H O ADP H+ TCA Cycle ATP O 2 2 Succinate FADH 2 Q IV H+ III H+ II I H+ NADH

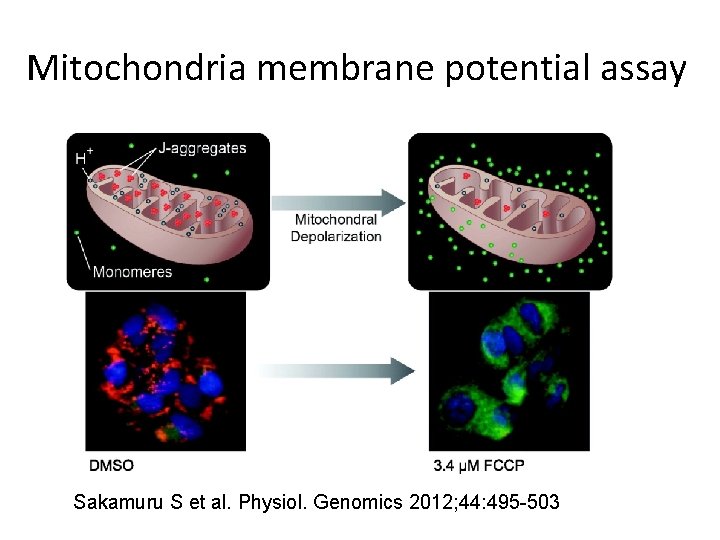
Mitochondria membrane potential assay Sakamuru S et al. Physiol. Genomics 2012; 44: 495 -503

Data Acquisition • Rat Oral and Intravenous LD 50 Data – – Public literature European Chemicals Agency (ECHA) e-Chem. Portal Chem. ID Plus • Fish 96 h LC 50 and daphnia 48 h LC 50 data – – EU Centre for Ecotox and Tox of Chemicals (ECETOC) Aquatic Japan US-EPA ecotoxicology DB from OECD QSAR Toolbox ECHA • Tox. Cast Mitochondrial toxicity data – – US-EPA Tox. Cast-II data download (v 12/13/2013) Name: Tox 21 Mitochondrial Toxicity Ratio Assessed in Hep. G 2 cells using Mito-MPS (BD Biosciences) AC 50 (in µM) was provided

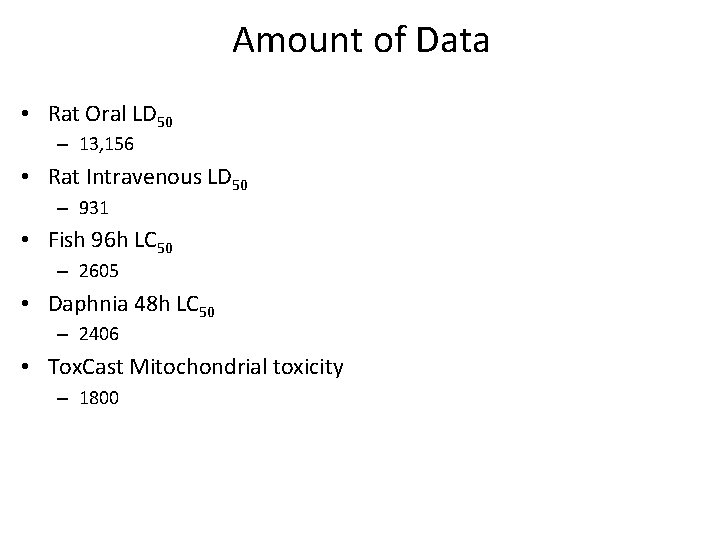
Amount of Data • Rat Oral LD 50 – 13, 156 • Rat Intravenous LD 50 – 931 • Fish 96 h LC 50 – 2605 • Daphnia 48 h LC 50 – 2406 • Tox. Cast Mitochondrial toxicity – 1800

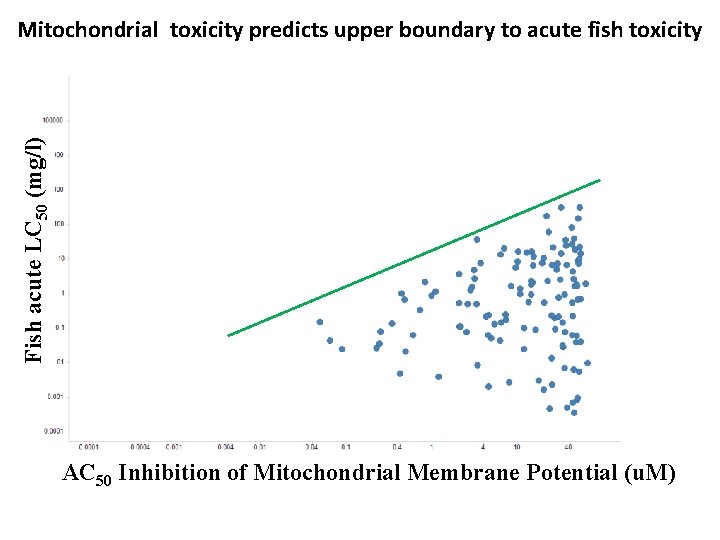
Mitochondrial toxicity predicts upper boundary to acute fish toxicity Fish acute LC 50 (mg/l) AC 50 Inhibition of Mitochondrial Membrane Potential ( AC 50 Inhibition of Mitochondrial Membrane Potential (u. M)

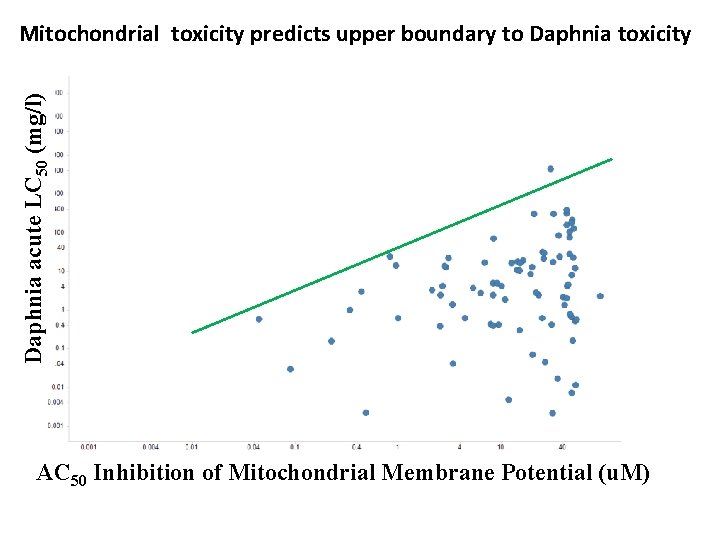
Mitochondrial toxicity predicts upper boundary to Daphnia toxicity Daphnia acute LC 50 (mg/l) AC 50 Inhibition of Mitochondrial Membrane Potential ( AC 50 Inhibition of Mitochondrial Membrane Potential (u. M)

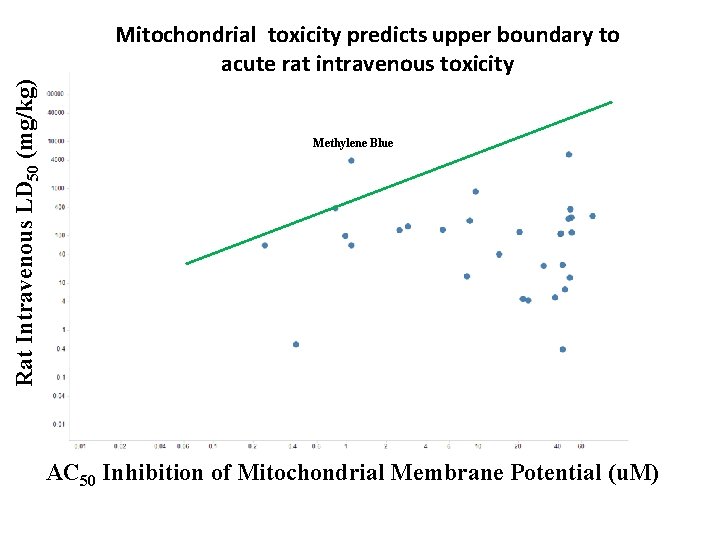
Rat Intravenous LD 50 (mg/kg) AC 50 Inhibition of Mitochondrial Membrane Potential ( Mitochondrial toxicity predicts upper boundary to acute rat intravenous toxicity AC 50 Inhibition of Mitochondri al Membrane Potential ( Methylene Blue AC 50 Inhibition of Mitochondrial Membrane Potential (u. M)

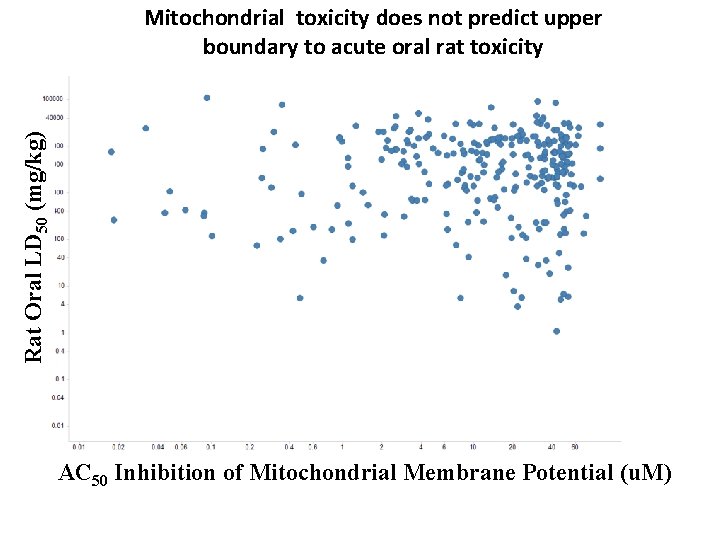
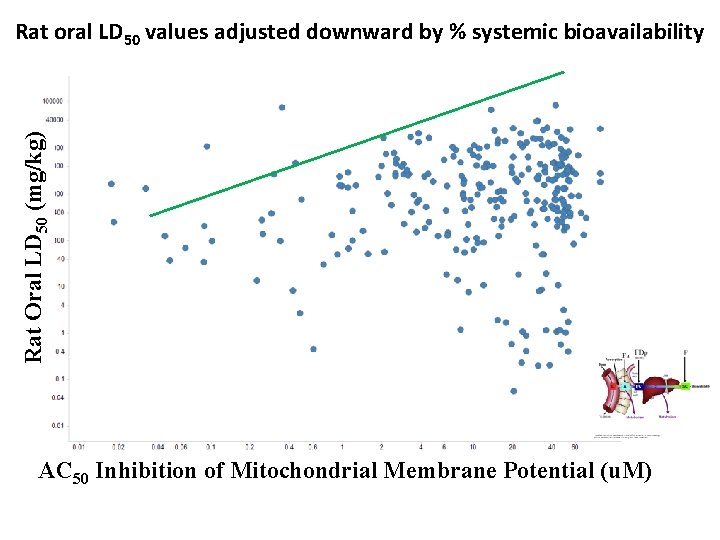
Rat Oral LD 50 (mg/kg) AC 50 Inhibition of Mitochondrial Membrane Potential ( Mitochondrial toxicity does not predict upper boundary to acute oral rat toxicity AC 50 Inhibition of Mitochondri al Membrane Potential ( AC 50 Inhibition of Mitochondrial Membrane Potential (u. M)

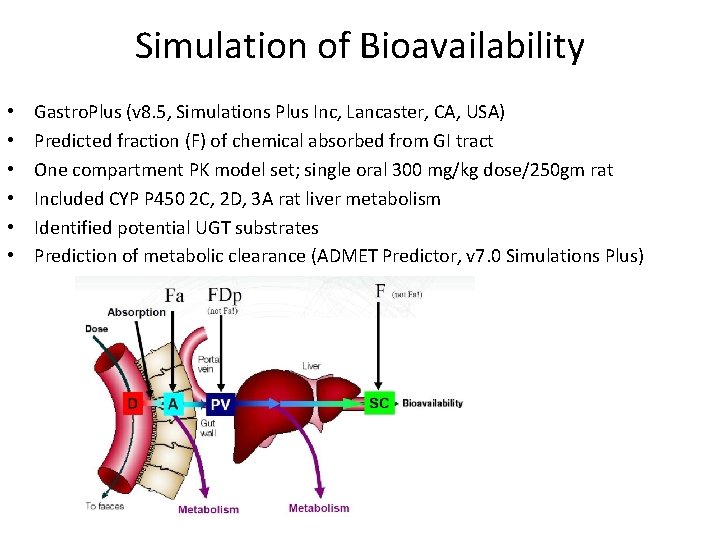
Simulation of Bioavailability • • • Gastro. Plus (v 8. 5, Simulations Plus Inc, Lancaster, CA, USA) Predicted fraction (F) of chemical absorbed from GI tract One compartment PK model set; single oral 300 mg/kg dose/250 gm rat Included CYP P 450 2 C, 2 D, 3 A rat liver metabolism Identified potential UGT substrates Prediction of metabolic clearance (ADMET Predictor, v 7. 0 Simulations Plus)

Rat oral LD 50 values adjusted downward by % systemic bioavailability Rat Oral LD 50 (mg/kg) AC 50 Inhibition of Mitochondrial Membrane Potential ( AC 50 Inhibition of Mitochondrial Membrane Potential (u. M)

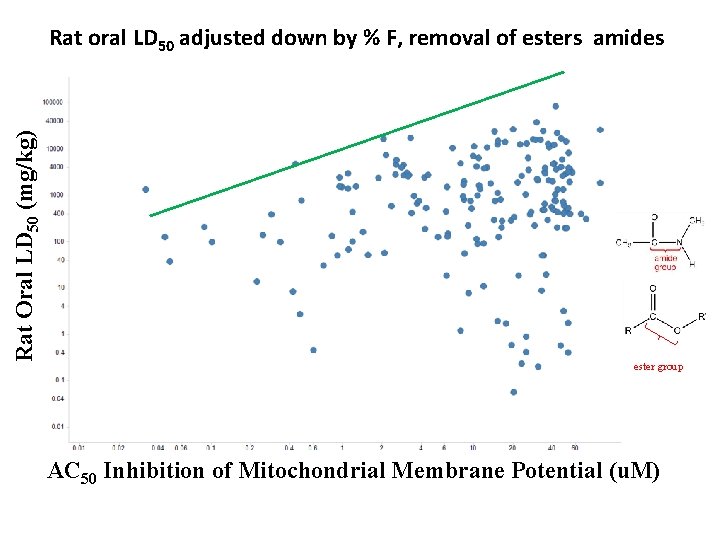
Rat oral LD 50 adjusted down by % F, removal of esters amides Rat Oral LD 50 (mg/kg) AC 50 Inhibition of Mitochondrial Membrane Potential ( AC 50 Inhibition of Mitochondri al Membrane Potential ( ester group AC 50 Inhibition of Mitochondrial Membrane Potential (u. M)

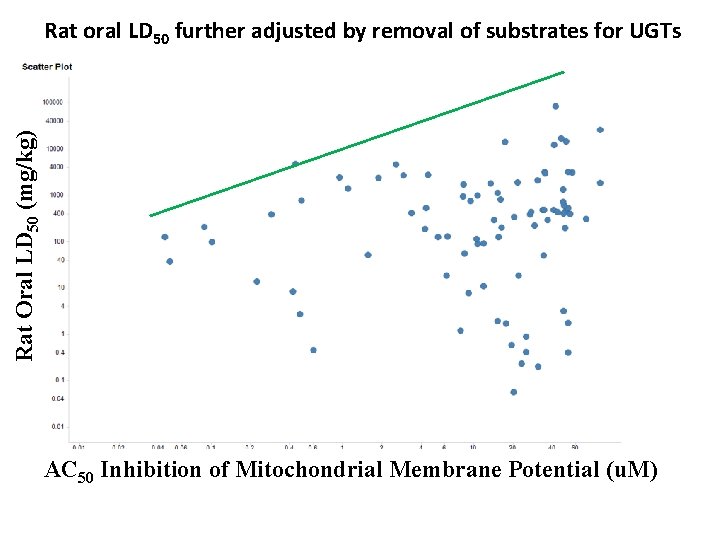
Rat Oral LD 50 (mg/kg) Rat oral LD 50 further adjusted by removal of substrates for UGTs AC 50 Inhibition of Mitochondri al Membrane Potential ( AC 50 Inhibition of Mitochondrial Membrane Potential (u. M)

Conclusions • HTS assays of mitochondrial toxicity can be used to predict high acute systemic toxicity in multiple species • Predictions of oral toxicity from HTS assays routes would often be confounded by chemical-specific differences in uptake and metabolism • With proper in silico estimations of absorption and first pass metabolism, HTS assays can eventually be used to predict acute oral toxicity in mammals • There is a need for in silico models that address hydrolysis • Other mechanistic endpoints need to be covered in vitro

Future opportunities: acute inhalation

Thank You! Questions?
 Tox cast
Tox cast Tox cast
Tox cast Ghs pictogram depicts which hazard
Ghs pictogram depicts which hazard Acute toxicity
Acute toxicity Acute toxicity
Acute toxicity Acute toxicity
Acute toxicity Shaped blockout
Shaped blockout B-tox peel before and after
B-tox peel before and after Comp tox
Comp tox Ideal mosfet
Ideal mosfet Efsa comprehensive european food consumption database
Efsa comprehensive european food consumption database Cox eox tox
Cox eox tox Word within the word list 14
Word within the word list 14 Christopher austin nih
Christopher austin nih Tox
Tox Poxymeter
Poxymeter B6 toxicity symptoms
B6 toxicity symptoms Mcgreafor
Mcgreafor Definition of chronic toxicity
Definition of chronic toxicity Vitamin d toxicity
Vitamin d toxicity Severe preeclampsia criteria
Severe preeclampsia criteria Symptoms of potassium
Symptoms of potassium What is oxygen toxicity
What is oxygen toxicity Organophosphate toxicity
Organophosphate toxicity Hepatic toxicity
Hepatic toxicity Lithium toxicity
Lithium toxicity Hypertiroidism
Hypertiroidism Definition of chronic toxicity
Definition of chronic toxicity Digoxin mechanism
Digoxin mechanism Kaexylate
Kaexylate Anticholinergic toxicity
Anticholinergic toxicity Alcohol toxicity
Alcohol toxicity Roles of vitamin c
Roles of vitamin c Digoxin toxicity symptoms
Digoxin toxicity symptoms Vitamin a functions
Vitamin a functions Treatment of salicylate toxicity
Treatment of salicylate toxicity Zinc toxicity
Zinc toxicity B complex action
B complex action Lithium toxicity
Lithium toxicity Toxicity index
Toxicity index Rais risk assessment
Rais risk assessment Mountains into molehills answer key
Mountains into molehills answer key Digoxin toxicity mnemonic
Digoxin toxicity mnemonic Dose response curve definition apes
Dose response curve definition apes Virtualization structure in cloud computing
Virtualization structure in cloud computing Mechanism and structure
Mechanism and structure Data organization and formatting of magnetic disk
Data organization and formatting of magnetic disk Basic mechanisms underlying seizures and epilepsy
Basic mechanisms underlying seizures and epilepsy Sensory and motor mechanisms
Sensory and motor mechanisms Security services of cryptography
Security services of cryptography Service delivery mechanism
Service delivery mechanism Security attacks services and mechanisms
Security attacks services and mechanisms Building vocabulary: diet categories and feeding mechanisms
Building vocabulary: diet categories and feeding mechanisms National mechanisms for reporting and follow-up
National mechanisms for reporting and follow-up Acute inflammation definition
Acute inflammation definition Classifying triangles 4-1
Classifying triangles 4-1 Cellular events of acute inflammation
Cellular events of acute inflammation Chapter 18 common chronic and acute conditions
Chapter 18 common chronic and acute conditions Morphological types of inflammation
Morphological types of inflammation Difference between acute and subacute rehab
Difference between acute and subacute rehab Differential diagnosis leukocytosis
Differential diagnosis leukocytosis Definition of periradicular disease
Definition of periradicular disease Using system.collections
Using system.collections Defrost using internal heat is accomplished using
Defrost using internal heat is accomplished using Mechanisms of movement lab report
Mechanisms of movement lab report What are freud's defense mechanisms
What are freud's defense mechanisms Countercurrent exchange thermoregulation
Countercurrent exchange thermoregulation Thermoneutral environment for neonates
Thermoneutral environment for neonates What are the ego defense mechanisms
What are the ego defense mechanisms Th and tc cells
Th and tc cells Reciprocal determinism psychology definition
Reciprocal determinism psychology definition Sponge defense mechanisms
Sponge defense mechanisms Automated scaling listener
Automated scaling listener