Knowledge Discovery in Microarray Gene Expression Data Gregory






- Slides: 41

Knowledge Discovery in Microarray Gene Expression Data Gregory Piatetsky-Shapiro gps@KDnuggets. com IMA 2002 Workshop on Data-driven Control and Optimization Copyright © 2002 KDnuggets

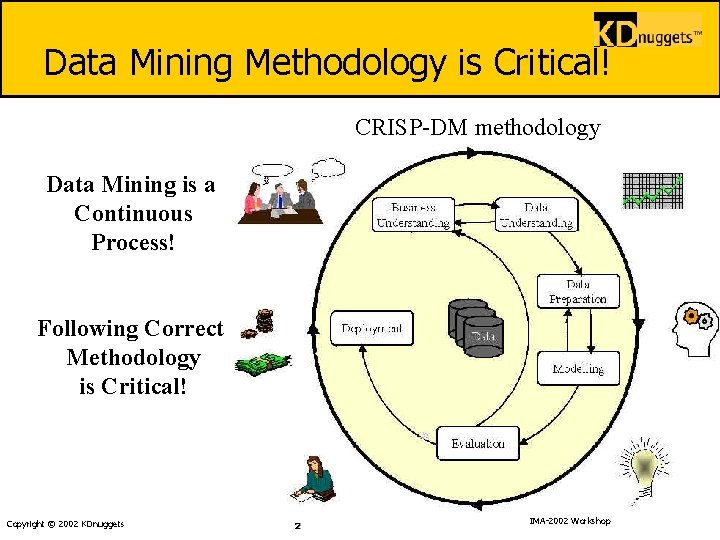
Data Mining Methodology is Critical! CRISP-DM methodology Data Mining is a Continuous Process! Following Correct Methodology is Critical! Copyright © 2002 KDnuggets 2 IMA-2002 Workshop

Overview § Molecular Biology Overview § Microarrays for Gene Expression § Classification on Microarray Data § avoiding false positives § wrapper approach § Microarrays for Modeling Dynamic Processes § finding causal networks and clusters Copyright © 2002 KDnuggets 3 IMA-2002 Workshop

Biology and Cells § All living organisms consist of cells. § Humans have trillions of cells. Yeast - one cell. § Cells are of many different types (blood, skin, nerve), but all arose from a single cell (the fertilized egg) § Each* cell contains a complete copy of the genome (the program for making the organism), encoded in DNA. Copyright © 2002 KDnuggets 4 IMA-2002 Workshop

DNA § DNA molecules are long double-stranded chains; 4 types of bases are attached to the backbone: adenine (A), guanine (G), cytosine (C), and thymine (T). A pairs with T, C with G. § A gene is a segment of DNA that specifies how to make a protein. § Human DNA has about 30 -35, 000 genes; Rice -- about 50 -60, 000, but shorter genes. Copyright © 2002 KDnuggets 5 IMA-2002 Workshop

Exons and Introns: Data and Logic? § exons are coding DNA (translated into a protein), which are only about 2% of human genome § introns are non-coding DNA, which provide structural integrity and regulatory (control) functions § exons can be thought of program data, while introns provide the program logic § Humans have much more control structure than rice Copyright © 2002 KDnuggets 6 IMA-2002 Workshop

Gene Expression § Cells are different because of differential gene expression. § About 40% of human genes are expressed at one time. § Gene is expressed by transcribing DNA into single-stranded m. RNA § m. RNA is later translated into a protein § Microarrays measure the level of m. RNA expression Copyright © 2002 KDnuggets 7 IMA-2002 Workshop

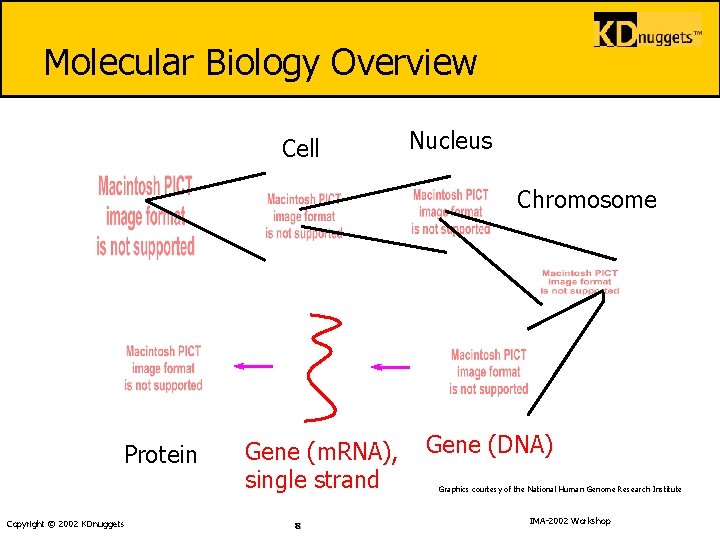
Molecular Biology Overview Cell Nucleus Chromosome Protein Copyright © 2002 KDnuggets Gene (m. RNA), single strand 8 Gene (DNA) Graphics courtesy of the National Human Genome Research Institute IMA-2002 Workshop

Gene Expression Measurement § m. RNA expression represents dynamic aspects of cell § m. RNA expression can be measured with latest technology § m. RNA is isolated and labeled with fluorescent protein § m. RNA is hybridized to the target; level of hybridization corresponds to light emission which is measured with a laser Copyright © 2002 KDnuggets 9 IMA-2002 Workshop

Gene Expression Microarrays The main types of gene expression microarrays: § Short oligonucleotide arrays (Affymetrix); § c. DNA or spotted arrays (Brown/Botstein). § Long oligonucleotide arrays (Agilent Inkjet); § Fiber-optic arrays §. . . Copyright © 2002 KDnuggets 10 IMA-2002 Workshop

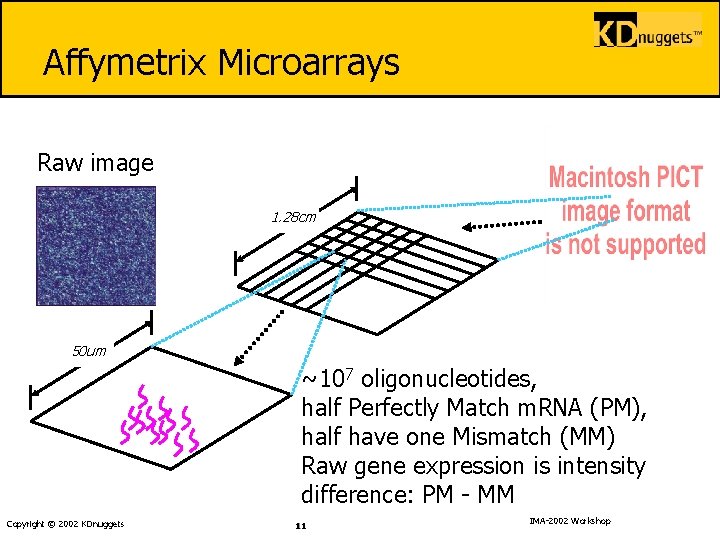
Affymetrix Microarrays Raw image 1. 28 cm 50 um ~107 oligonucleotides, half Perfectly Match m. RNA (PM), half have one Mismatch (MM) Raw gene expression is intensity difference: PM - MM Copyright © 2002 KDnuggets 11 IMA-2002 Workshop

Microarray Potential Applications § Biological discovery § new and better molecular diagnostics § new molecular targets for therapy § finding and refining biological pathways § Recent examples § molecular diagnosis of leukemia, breast cancer, . . . § appropriate treatment for genetic signature § potential new drug targets Copyright © 2002 KDnuggets 12 IMA-2002 Workshop

Microarray Data Analysis Types § Gene Selection § find genes for therapeutic targets § avoid false positives (FDA approval ? ) § Classification (Supervised) § identify disease § predict outcome / select best treatment § Clustering (Unsupervised) § find new biological classes / refine existing ones § exploration Copyright © 2002 KDnuggets 13 IMA-2002 Workshop

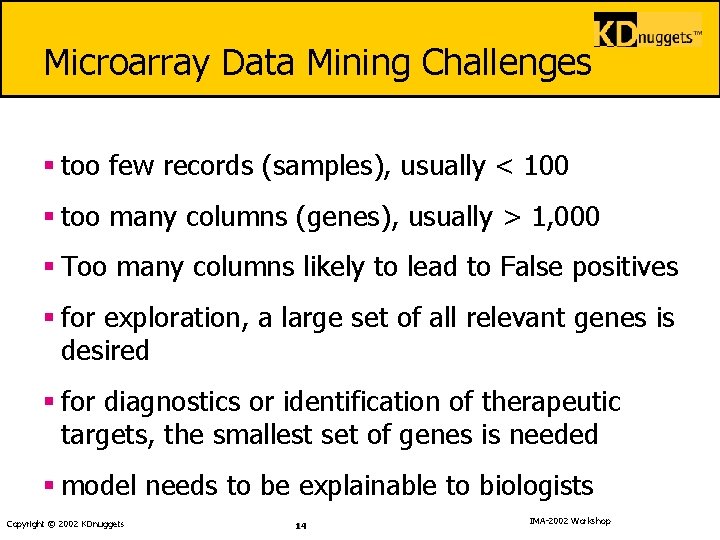
Microarray Data Mining Challenges § too few records (samples), usually < 100 § too many columns (genes), usually > 1, 000 § Too many columns likely to lead to False positives § for exploration, a large set of all relevant genes is desired § for diagnostics or identification of therapeutic targets, the smallest set of genes is needed § model needs to be explainable to biologists Copyright © 2002 KDnuggets 14 IMA-2002 Workshop

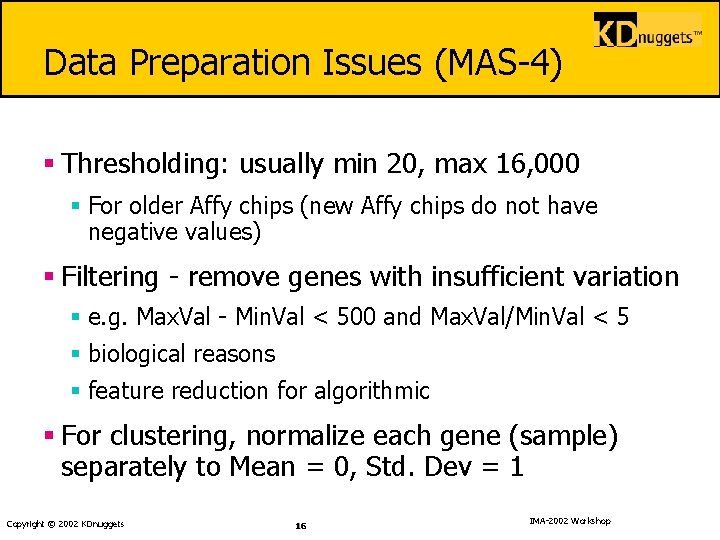
Data Preparation Issues (MAS-4) § Thresholding: usually min 20, max 16, 000 § For older Affy chips (new Affy chips do not have negative values) § Filtering - remove genes with insufficient variation § e. g. Max. Val - Min. Val < 500 and Max. Val/Min. Val < 5 § biological reasons § feature reduction for algorithmic § For clustering, normalize each gene (sample) separately to Mean = 0, Std. Dev = 1 Copyright © 2002 KDnuggets 16 IMA-2002 Workshop

Classification § desired features: § robust in presence of false positives § understandable § return confidence/probability § fast enough § simplest approaches are most robust § advanced approaches can be more accurate Copyright © 2002 KDnuggets 17 IMA-2002 Workshop

FALSE POSITIVES PROBLEM § Not enough records (samples), usually < 100 § Too many columns (genes), usually >>1, 000 § FALSE POSITIVES are very likely because of few records and many columns Copyright © 2002 KDnuggets 18 IMA-2002 Workshop

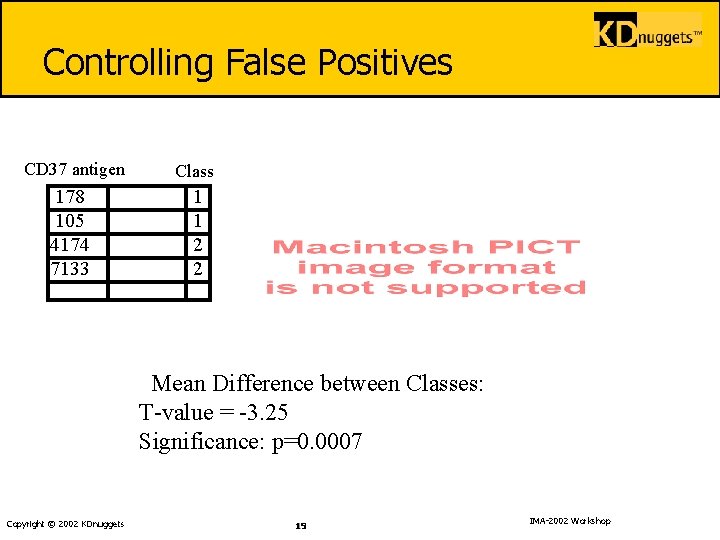
Controlling False Positives CD 37 antigen Class 178 105 4174 7133 1 1 2 2 Mean Difference between Classes: T-value = -3. 25 Significance: p=0. 0007 Copyright © 2002 KDnuggets 19 IMA-2002 Workshop

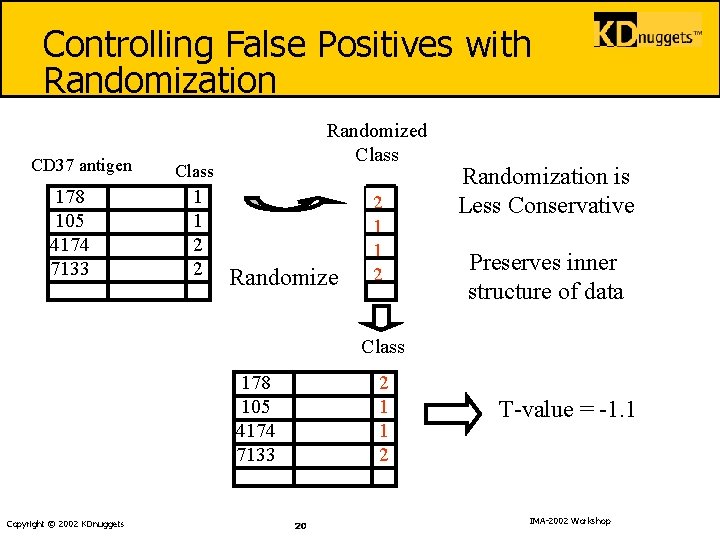
Controlling False Positives with Randomization CD 37 antigen 178 105 4174 7133 Randomized Class 1 1 2 2 Randomize 2 1 1 2 Randomization is Less Conservative Preserves inner structure of data Class 178 105 4174 7133 Copyright © 2002 KDnuggets 2 1 1 2 20 T-value = -1. 1 IMA-2002 Workshop

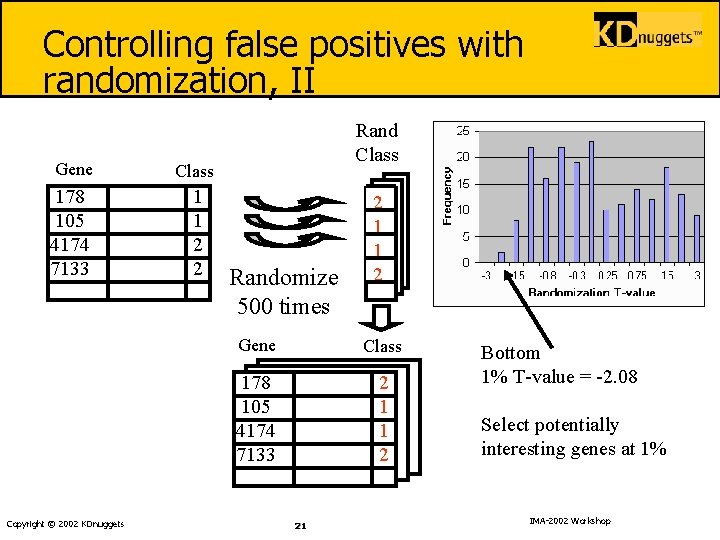
Controlling false positives with randomization, II Gene Class 178 105 4174 7133 1 1 2 2 Copyright © 2002 KDnuggets Rand Class Randomize 500 times 2 1 1 2 Gene Class 178 105 4174 7133 2 1 1 2 21 Bottom 1% T-value = -2. 08 Select potentially interesting genes at 1% IMA-2002 Workshop

Controlling False Positives: SAM (Statistical Analysis of Microarrays) § Tusher, Tibshirani, and Chu, Significance analysis of microarrays …, PNAS, Apr 2001 § SAM software available from Tibshirani web site Copyright © 2002 KDnuggets 22 IMA-2002 Workshop

Feature selection approach § Rank genes by measure; select top 200 -500 § T-test for Mean Difference= § Signal to Noise (S 2 N) = § Other: Information-based, biological? § Almost any method works well with a good feature selection Copyright © 2002 KDnuggets 24 IMA-2002 Workshop

Gene Reduction improves Classification § most learning algorithms looks for non-linear combinations of features -- can easily find many spurious combinations given small # of records and large # of genes § Classification accuracy improves if we first reduce # of genes by a linear method, e. g. T-values of mean difference § Heuristic: select equal # genes from each class § Then apply a favorite machine learning algorithm Copyright © 2002 KDnuggets 25 IMA-2002 Workshop

Wrapper approach to select the best gene set Select best 200 or so genes based on statistical measures Test models using 1, 2, 3, …, 10, 20, 30, 40, . . . genes with xvalidation. Select gene set with lowest average error Heuristically, at least 10 genes overall Copyright © 2002 KDnuggets 26 IMA-2002 Workshop

Popular Classification Methods § Decision Trees/Rules § find smallest gene sets, but not robust false positives § Neural Nets - work well for reduced # of genes § K-nearest neighbor - robust for small # genes § Tree. Net from authors of CART and MARS § networks of simple trees; very robust against outliers § Support Vector Machines (SVM) § good accuracy, does its own gene selection, but hard to understand §. . . Copyright © 2002 KDnuggets 27 IMA-2002 Workshop

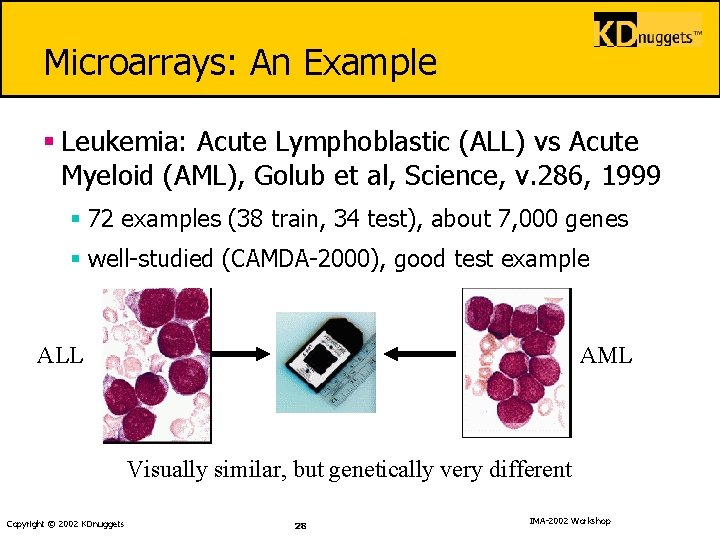
Microarrays: An Example § Leukemia: Acute Lymphoblastic (ALL) vs Acute Myeloid (AML), Golub et al, Science, v. 286, 1999 § 72 examples (38 train, 34 test), about 7, 000 genes § well-studied (CAMDA-2000), good test example ALL AML Visually similar, but genetically very different Copyright © 2002 KDnuggets 28 IMA-2002 Workshop

Results on the test data § Genes selected and model trained on Train set ONLY! § Best Clementine neural net model used 10 genes per class § Evaluation on test data (34 samples) gives § 1 or 2 errors (94 -97% accuracy), § Note: all methods give error on sample 66, believed to be mis-classified by a pathologist Copyright © 2002 KDnuggets 29 IMA-2002 Workshop

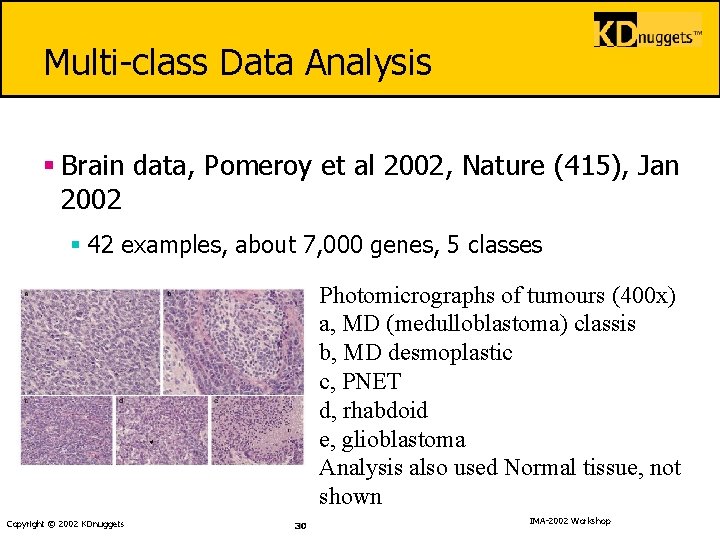
Multi-class Data Analysis § Brain data, Pomeroy et al 2002, Nature (415), Jan 2002 § 42 examples, about 7, 000 genes, 5 classes Photomicrographs of tumours (400 x) a, MD (medulloblastoma) classis b, MD desmoplastic c, PNET d, rhabdoid e, glioblastoma Analysis also used Normal tissue, not shown Copyright © 2002 KDnuggets 30 IMA-2002 Workshop

Modeling with Tree. Net § Build a model using top 3 genes from each class § Evaluate using cross-validation § Results: 95% accuracy: § 1 error on training data, 1 on test Copyright © 2002 KDnuggets 31 IMA-2002 Workshop

Tree. Net results for multi-class data Average cross-validation accuracy over 95% Original authors had accuracy of about 85% using nearest neighbor classifier. Copyright © 2002 KDnuggets 32 IMA-2002 Workshop

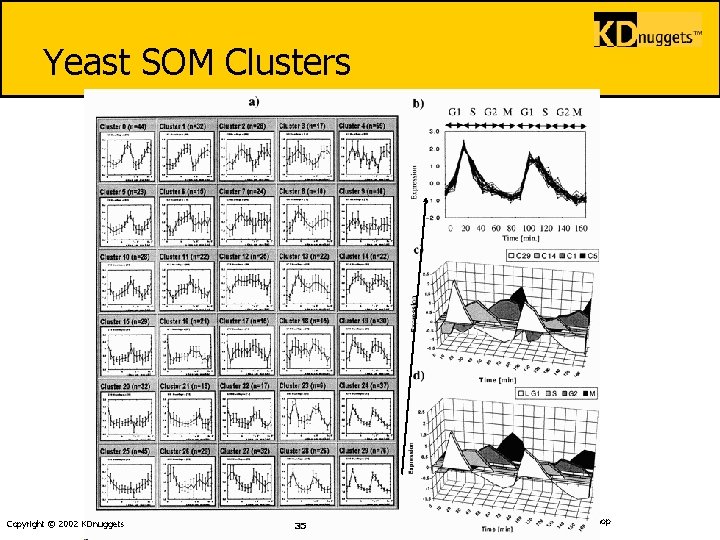
Yeast SOM Clusters § Yeast Cell Cycle SOM. www. pnas. org/cgi/content/full/96/6/2907 § (a) 6 × 5 SOM. The 828 genes that passed the variation filter were grouped into 30 clusters. Each cluster is represented by the centroid (average pattern) for genes in the cluster. Expression level of each gene was normalized to have mean = 0 and SD = 1 across time points. Expression levels are shown on y-axis and time points on x-axis. Error bars indicate the SD of average expression. n indicates the number of genes within each cluster. Note that multiple clusters exhibit periodic behavior and that adjacent clusters have similar behavior. (b) Cluster 29 detail. Cluster 29 contains 76 genes exhibiting periodic behavior with peak expression in late G 1. Normalized expression pattern of 30 genes nearest the centroid are shown. (c) Centroids for SOMderived clusters 29, 14, 1, and 5, corresponding to G 1, S, G 2 and M phases of the cell cycle, are shown. Copyright © 2002 KDnuggets 34 IMA-2002 Workshop

Yeast SOM Clusters Copyright © 2002 KDnuggets 35 IMA-2002 Workshop

Discovery of causal processes § A long term goal of Systems Biology is to discover the causal processes among genes, proteins, and other molecules in cells § Can this be done (in part) by using data from High Throughput experiments, such as microarrays? Copyright © 2002 KDnuggets 36 IMA-2002 Workshop

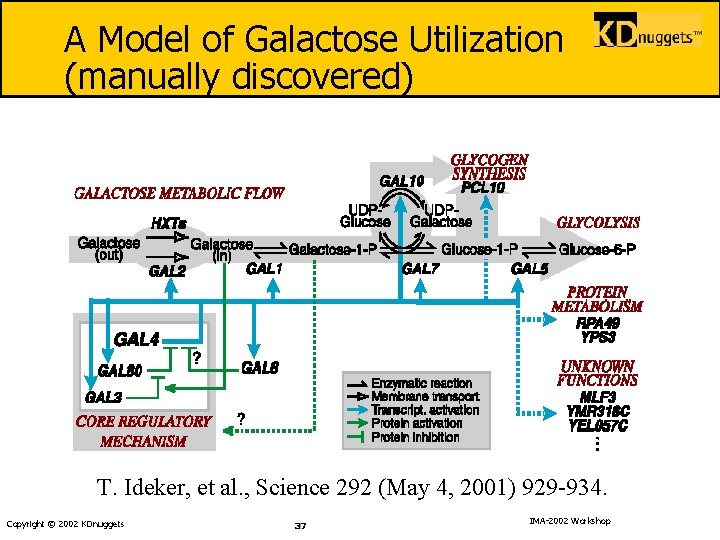
A Model of Galactose Utilization (manually discovered) T. Ideker, et al. , Science 292 (May 4, 2001) 929 -934. Copyright © 2002 KDnuggets 37 IMA-2002 Workshop

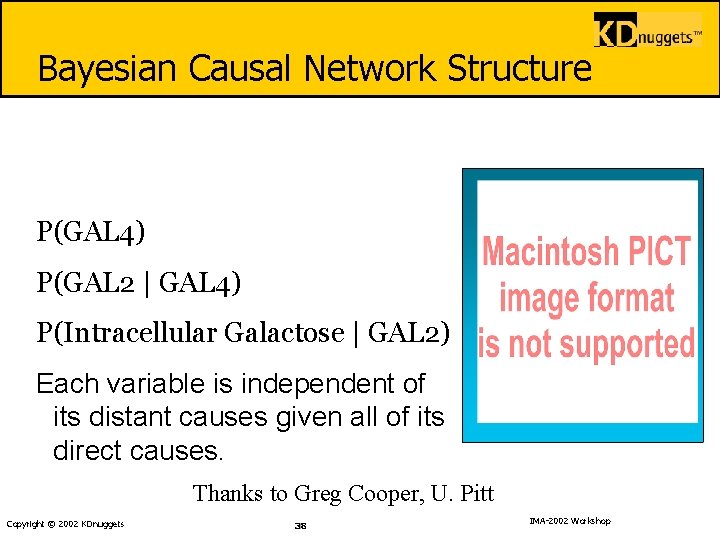
Bayesian Causal Network Structure P(GAL 4) P(GAL 2 | GAL 4) P(Intracellular Galactose | GAL 2) Each variable is independent of its distant causes given all of its direct causes. Thanks to Greg Cooper, U. Pitt Copyright © 2002 KDnuggets 38 IMA-2002 Workshop

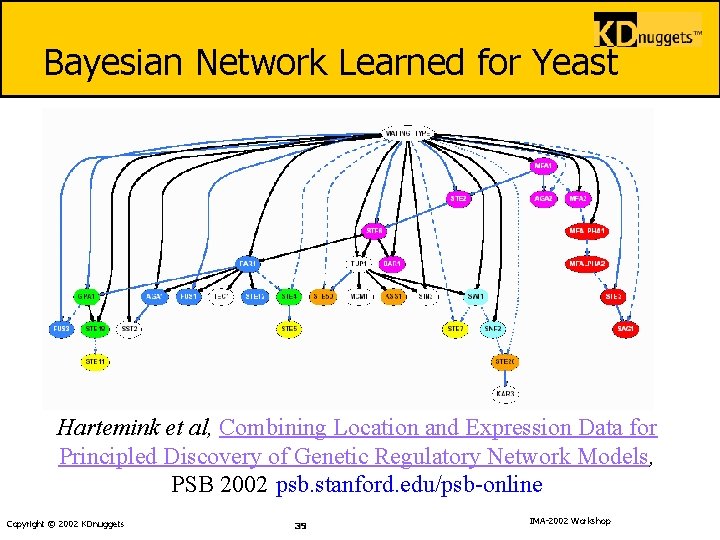
Bayesian Network Learned for Yeast Hartemink et al, Combining Location and Expression Data for Principled Discovery of Genetic Regulatory Network Models, PSB 2002 psb. stanford. edu/psb-online Copyright © 2002 KDnuggets 39 IMA-2002 Workshop

Future directions for Microarray Analysis § Algorithms optimized for small samples § Integration with other data § biological networks § medical text § protein data § Cost-sensitive classification algorithms § error cost depends on outcome (don’t want to miss treatable cancer), treatment side effects, etc. Copyright © 2002 KDnuggets 40 IMA-2002 Workshop

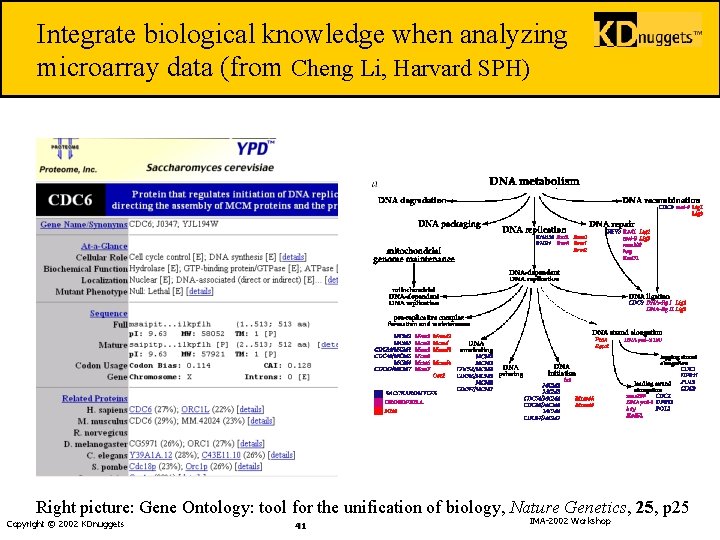
Integrate biological knowledge when analyzing microarray data (from Cheng Li, Harvard SPH) Right picture: Gene Ontology: tool for the unification of biology, Nature Genetics, 25, p 25 Copyright © 2002 KDnuggets 41 IMA-2002 Workshop

Gene. Spring Demo § Yeast data § Zoom all the way to bases § Yeast Cycle -- animation § Color -- expression strength Copyright © 2002 KDnuggets 42 IMA-2002 Workshop

Acknowledgements § Sridhar Ramaswamy, MIT Whitehead Institute § Pablo Tamayo, MIT Whitehead Institute § Greg Cooper, U. Pittsburgh § Tom Khabaza, SPSS Copyright © 2002 KDnuggets 43 IMA-2002 Workshop

Thank you! Further resources on Data Mining: www. KDnuggets. com Contact: Gregory Piatetsky-Shapiro: gps@KDnuggets. com Copyright © 2002 KDnuggets 44 IMA-2002 Workshop