Functional Annotation of the Horse Genomic Annotation and








- Slides: 29

Functional Annotation of the Horse Genomic Annotation and Functional Modeling Workshop Maxwell H. Gluck Equine Research Center 15 -16 November, 2011

Genomic Annotation p 1. 2. p Genome annotation is the process of attaching biological information to genomic sequences. It consists of two key steps: identifying functional elements in the genome: “structural annotation” attaching biological information to these elements: “functional annotation” biologists often use the term “annotation” when they are referring only to structural annotation

Structural & Functional Annotation Structural Annotation: p Open reading frames (ORFs) predicted during genome assembly p predicted ORFs require experimental confirmation Functional Annotation: p initially, predicted ORFs have no functional literature and functional annotation relies on sequence analysis, etc p functional literature exists for many genes/proteins prior to genome sequencing p functional annotation does not rely on a completed genome sequence! n relies on molecular data

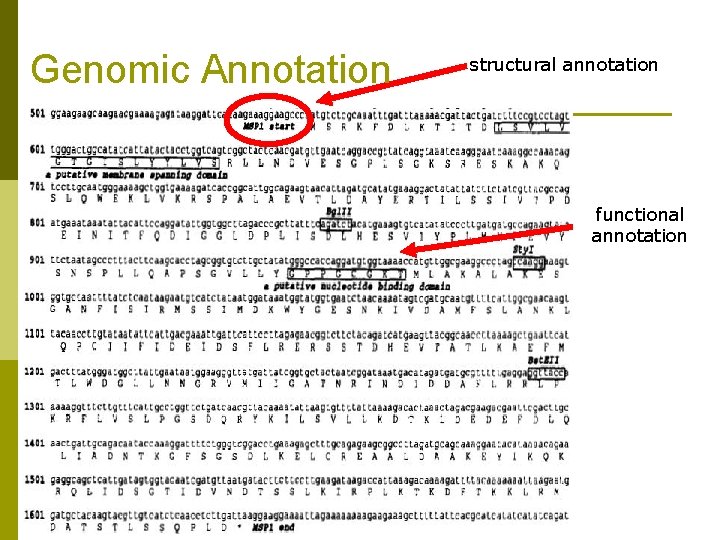
Genomic Annotation structural annotation functional annotation

Genomic Annotation Genomic annotation is distributed across different databases. p Databases: have different file formats, annotation pipelines, etc. p Biologists: don’t care (just want their data…) p Problem: how to exchange/share genomic annotations from different databases? p

Bio-ontologies Ontologies first used in biology to enable databases to share & exchange data. p Bio-ontologies are used to capture biological information in a way that can be read by both humans and computers p n n p annotate data in a consistent way allows data sharing across databases These same features allow computational analysis of high-throughput “omics” datasets using ontologies.

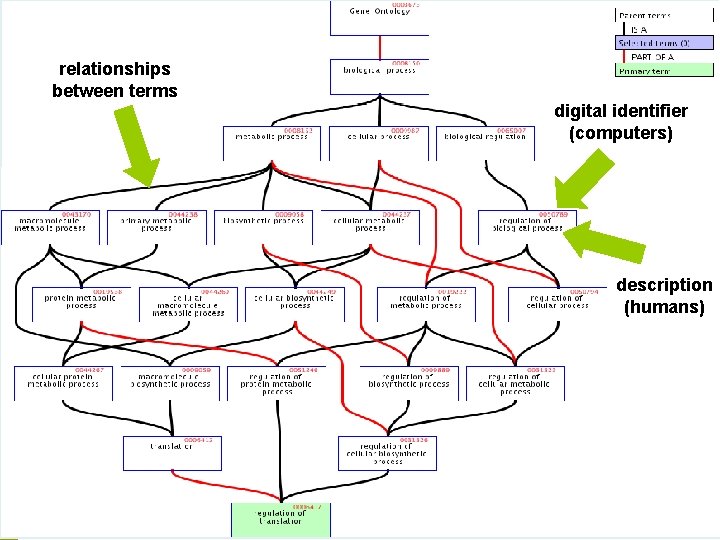
relationships between terms Ontologies digital identifier (computers) description (humans)

Ontologies & Genomic Annotation Structural Annotation: p Open reading frames (ORFs) predicted during genome assembly p predicted ORFs require experimental confirmation p Sequence Ontology Project (SO): provide for a structured controlled vocabulary for the description of primary annotations of nucleic acid sequence Functional Annotation: p initially, predicted ORFs have no functional literature and GO annotation relies on computational methods (rapid) p functional literature exists for many genes/proteins prior to genome sequencing p Gene Ontology (GO): annotation of gene product function

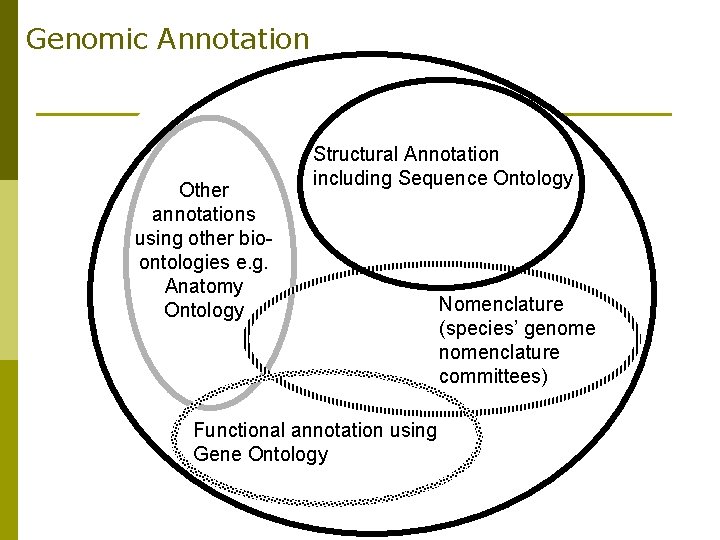
Genomic Annotation Other annotations using other bioontologies e. g. Anatomy Ontology Structural Annotation including Sequence Ontology Functional annotation using Gene Ontology Nomenclature (species’ genomenclature committees)

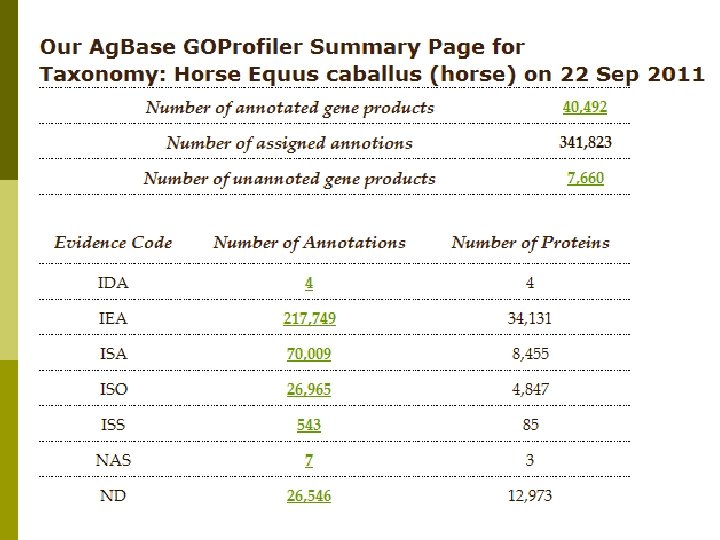
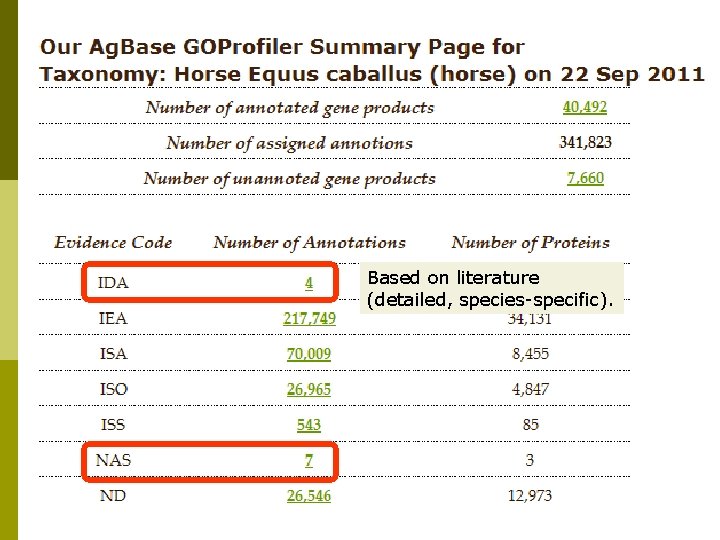
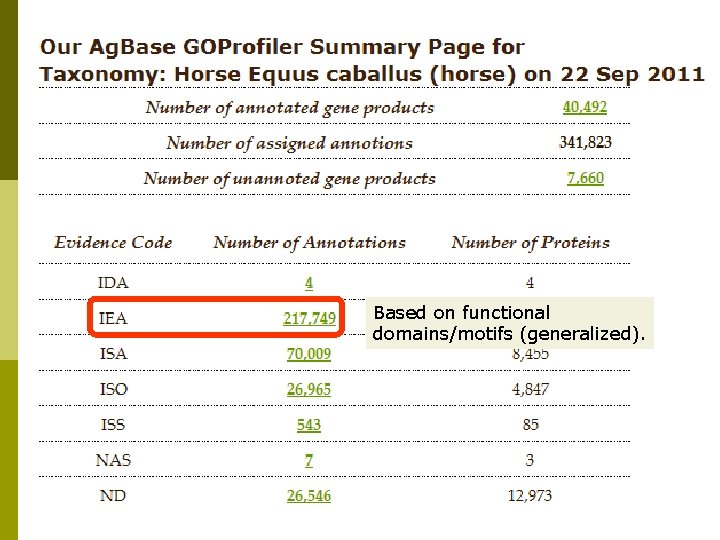
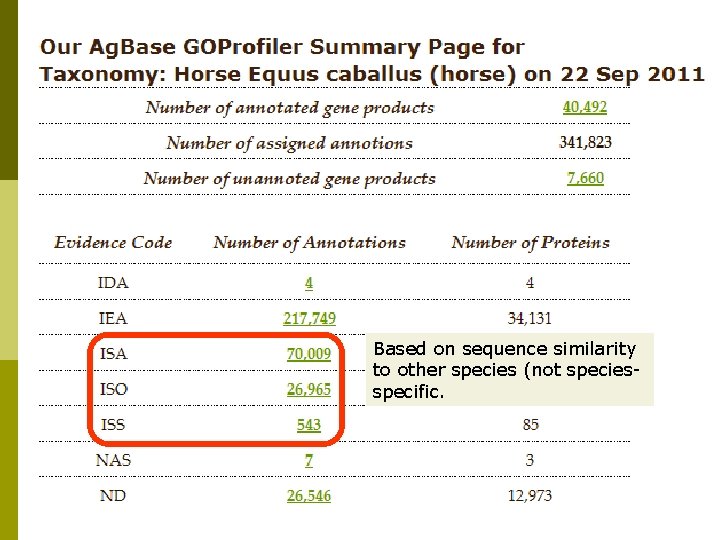
Horse GO Annotation p p EBI GOA provides sequence-based GO annotation for all Uni. Prot. KB records based upon analysis of functional motifs and domains. Ag. Base provides sequence-based GO for horse protein records not in Uni. Prot. KB (e. g. NCBI predicted proteins). Ag. Base also provides sequence-based GO annotation for ESTs, m. RNAs with no linked protein records if they are linked on a microarray (or by request). For gene products with published literature, Ag. Base provides experimental based GO annotation.

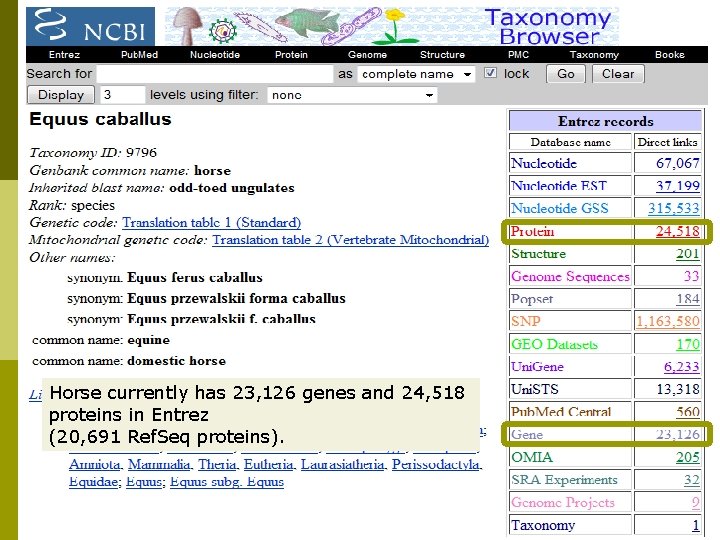
Horse currently has 23, 126 genes and 24, 518 proteins in Entrez (20, 691 Ref. Seq proteins).

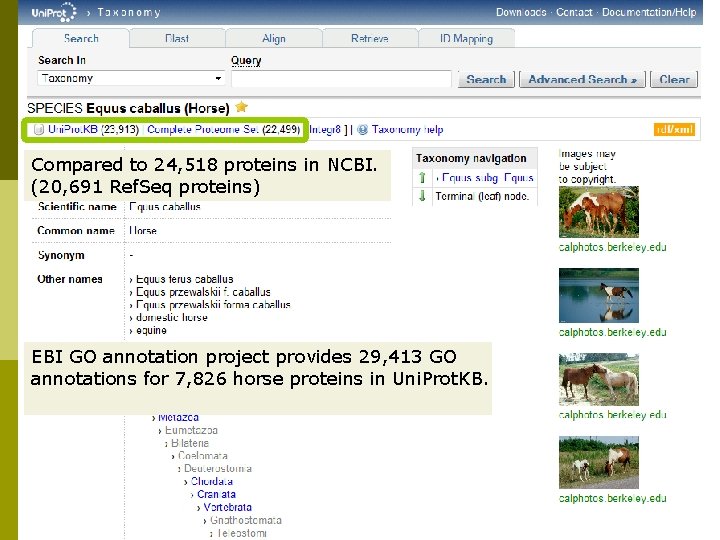
Compared to 24, 518 proteins in NCBI. (20, 691 Ref. Seq proteins) EBI GO annotation project provides 29, 413 GO annotations for 7, 826 horse proteins in Uni. Prot. KB.

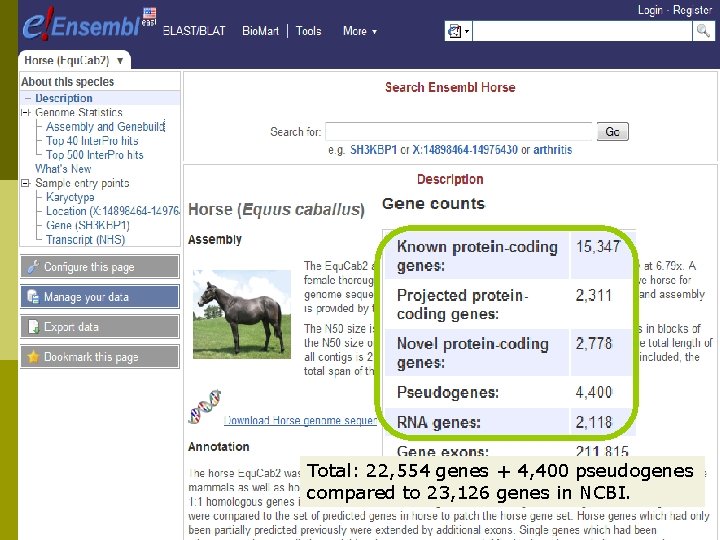
Total: 22, 554 genes + 4, 400 pseudogenes compared to 23, 126 genes in NCBI.

Horse Genes unique to NCBI (3, 955) genes with overlap (19, 462) Genes unique to Ensembl (26, 954) These numbers based upon published gene numbers from both NCBI & Ensembl. How important is it for the community to have a reference gene set?

1. Provide annotations (data) 2. Provide tools (to help with modeling using this data) 3. Provide training (to enable use of the data & tools)


Based on literature (detailed, species-specific).

Based on functional domains/motifs (generalized).

Based on sequence similarity to other species (not speciesspecific.

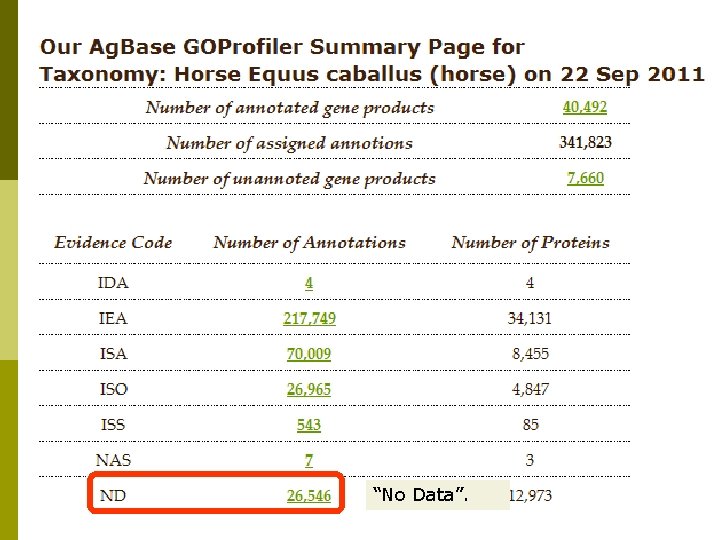
“No Data”.

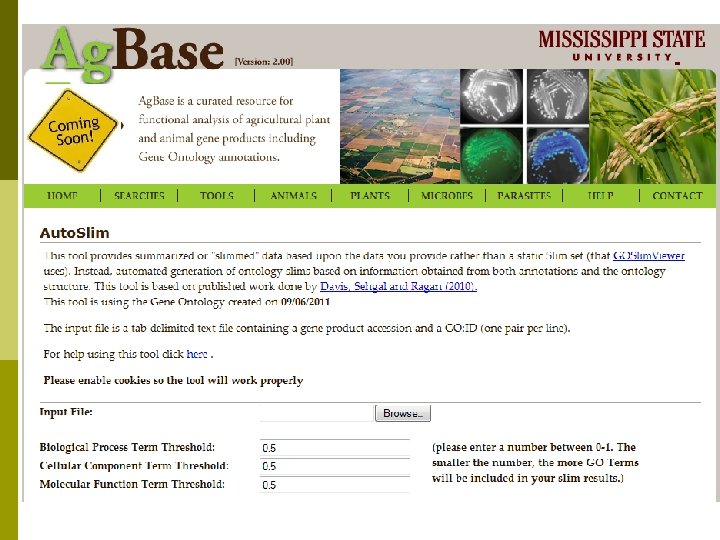
Ag. Base: the next three years p New grant will expand biocuration effort n n p Provide tools and HPC support for larger data sets. n n n p Biocuration for USDA Animal Genome species. Capture functional information in other ontologies (e. g. , anatomy, disease cell type). Focus on genes that have no literature. Continued funding for workshops. Helping by request – bioinformatics, biocomputing, functional modeling. Linking phenotype to traits and functions. n n Comparative browsers Tools to help locate QTL candidate genes. PROJ NO: MIS-391110 AGENCY

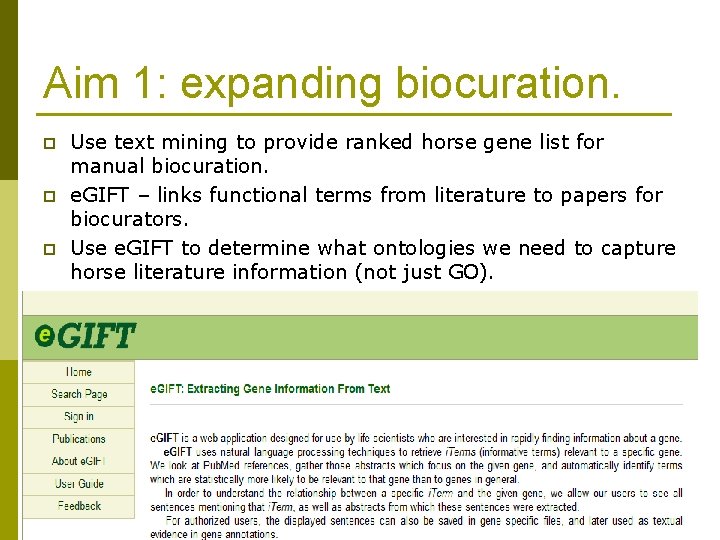
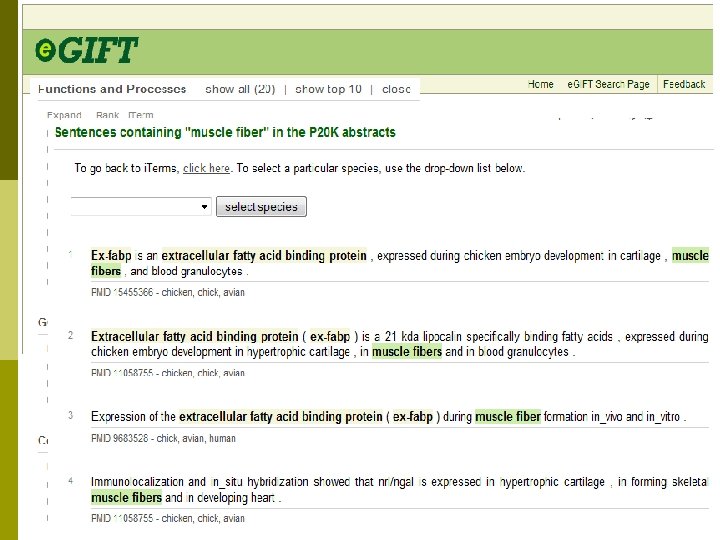
Aim 1: expanding biocuration. p p p Use text mining to provide ranked horse gene list for manual biocuration. e. GIFT – links functional terms from literature to papers for biocurators. Use e. GIFT to determine what ontologies we need to capture horse literature information (not just GO).


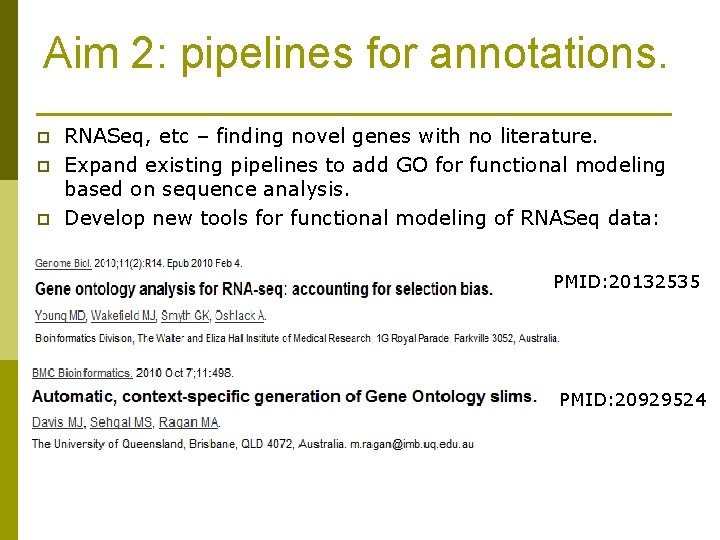
Aim 2: pipelines for annotations. p p p RNASeq, etc – finding novel genes with no literature. Expand existing pipelines to add GO for functional modeling based on sequence analysis. Develop new tools for functional modeling of RNASeq data: PMID: 20132535 PMID: 20929524


Aim 3: Linking genotype to phenotype Provide comparative genome browsers to enable cross-species comparison of common traits. p Phenotype – anatomy, physiology, behavior. p n n Working with NSF funded groups to develop phenotype annotations. Use data to link phenotype to genotype.

Horse Genome Annotation Ag. Base will provide ranked gene list for horse – community feedback and comment. p User requests for annotation also. p Continuing support: bioinformatics, biocomputing, tool development, help with functional modeling. p What do you want? p

Workshop p This afternoon: n n p Tomorrow (hands on): n n n p more detail about functional modeling a working knowledge of functional annotation (GO) adding GO to a data set mapping accessions working on your own data sets example data to work on tutorial – learning more about GO WIIFM – making connections between molecular and ‘omics’; back to biology

 David annotation tool
David annotation tool Genomic equivalence definition
Genomic equivalence definition Genomic england
Genomic england Genomic england
Genomic england Anneke seller
Anneke seller Genomic instability
Genomic instability Genomic
Genomic Genomic imprinting definition
Genomic imprinting definition Genomic signal processing
Genomic signal processing Comparative genomic hybridization animation
Comparative genomic hybridization animation Genomic equivalence definition
Genomic equivalence definition Space maintainer definition
Space maintainer definition Non functional plasma enzymes
Non functional plasma enzymes Enzymes of blood plasma
Enzymes of blood plasma Functional and non functional
Functional and non functional Sequence assembly ppt
Sequence assembly ppt Hát kết hợp bộ gõ cơ thể
Hát kết hợp bộ gõ cơ thể Slidetodoc
Slidetodoc Bổ thể
Bổ thể Tỉ lệ cơ thể trẻ em
Tỉ lệ cơ thể trẻ em Voi kéo gỗ như thế nào
Voi kéo gỗ như thế nào Thang điểm glasgow
Thang điểm glasgow Hát lên người ơi
Hát lên người ơi Môn thể thao bắt đầu bằng từ chạy
Môn thể thao bắt đầu bằng từ chạy Thế nào là hệ số cao nhất
Thế nào là hệ số cao nhất Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Cong thức tính động năng
Cong thức tính động năng Trời xanh đây là của chúng ta thể thơ
Trời xanh đây là của chúng ta thể thơ Mật thư anh em như thể tay chân
Mật thư anh em như thể tay chân Làm thế nào để 102-1=99
Làm thế nào để 102-1=99