Computational Analysis of the Taxanomical Classification of Short




- Slides: 22

Computational Analysis of the Taxanomical Classification of Short 16 S r. RNA Sequences Christel Chehoud Mentor: Brian Haas

Overview l l l Human Microbiome Project 16 S r. RNA Reference and Test Sets Classifiers Accuracy of Classifications Results

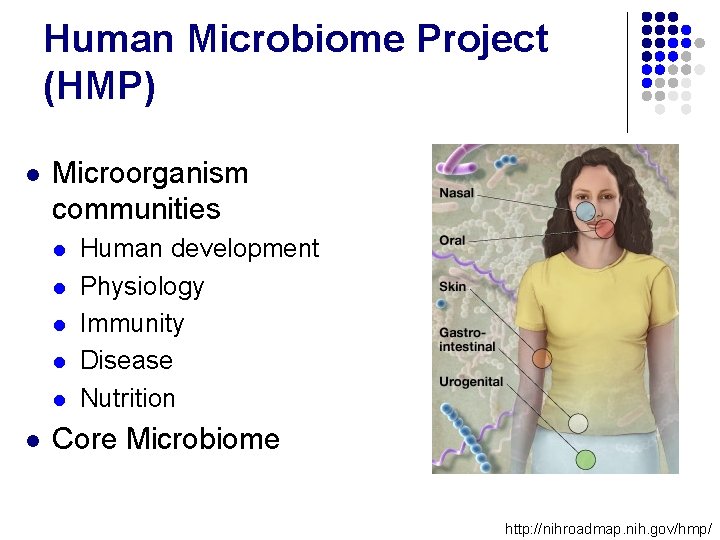
Human Microbiome Project (HMP) l Microorganism communities l l l Human development Physiology Immunity Disease Nutrition Core Microbiome http: //nihroadmap. nih. gov/hmp/

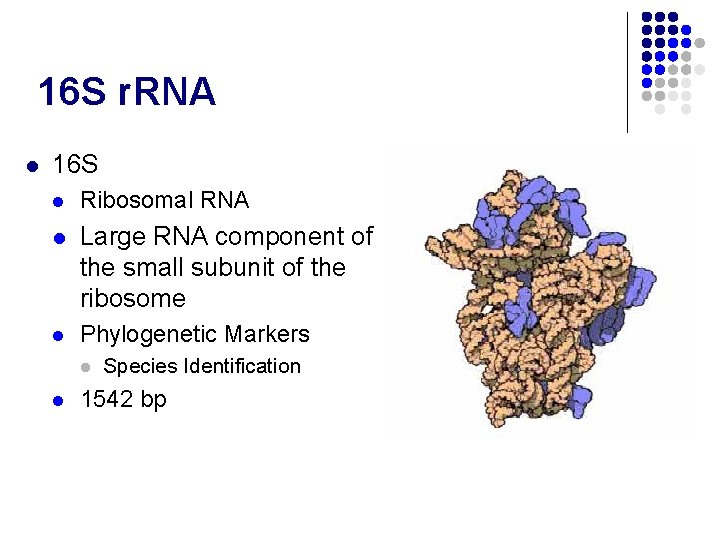
16 S r. RNA l 16 S l Ribosomal RNA l Large RNA component of the small subunit of the ribosome l Phylogenetic Markers l l Species Identification 1542 bp

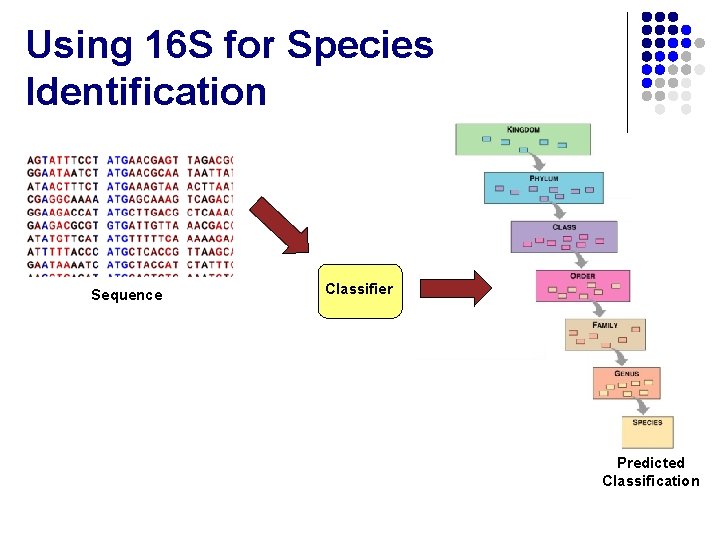
Using 16 S for Species Identification Sequence Classifier Predicted Classification

Project Goal l l New Sequencing Technology Evaluate the accuracy of the classification of the 16 S r. RNA across different: l l l Classifiers Regions of the sequence Phylogeny

Reference Dataset l RDP Core Set l l l l Trusted Taxonomies 6, 621 sequences Phylum: 27 Class: 43 Order: 97 Family: 258 Genus: 1352

Green. Genes’s Full Collection of Sequences l l Full Collection used by Green. Genes High phylogenetic diversity l 188, 073 sequences

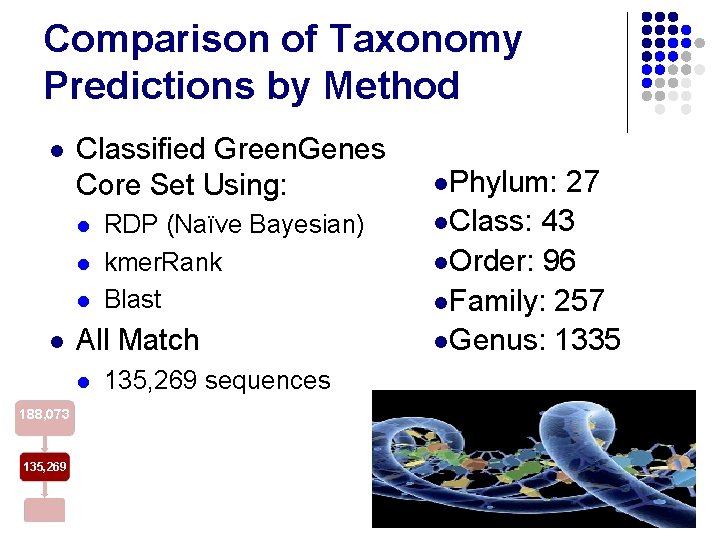
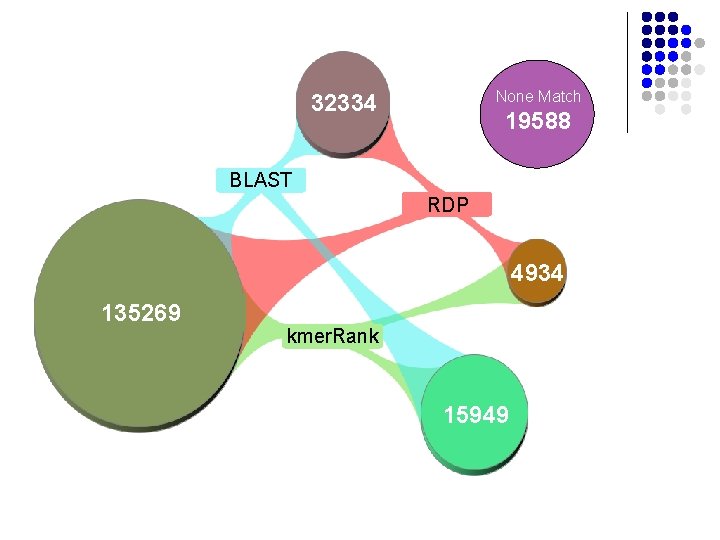
Comparison of Taxonomy Predictions by Method l Classified Green. Genes Core Set Using: l l All Match l 188, 073 135, 269 RDP (Naïve Bayesian) kmer. Rank Blast 135, 269 sequences l. Phylum: 27 l. Class: 43 l. Order: 96 l. Family: 257 l. Genus: 1335

None Match: 19588 None Match 32334 19588 BLAST RDP 4934 135269 kmer. Rank 15949

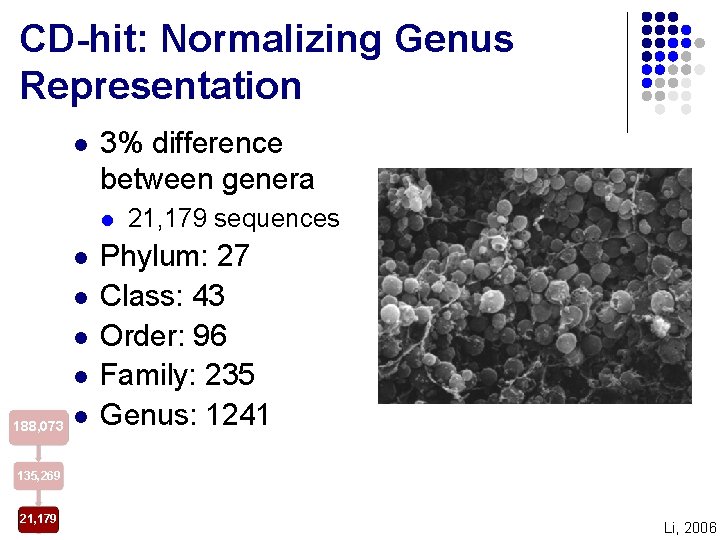
CD-hit: Normalizing Genus Representation l 3% difference between genera l l l 188, 073 l 21, 179 sequences Phylum: 27 Class: 43 Order: 96 Family: 235 Genus: 1241 135, 269 21, 179 Li, 2006

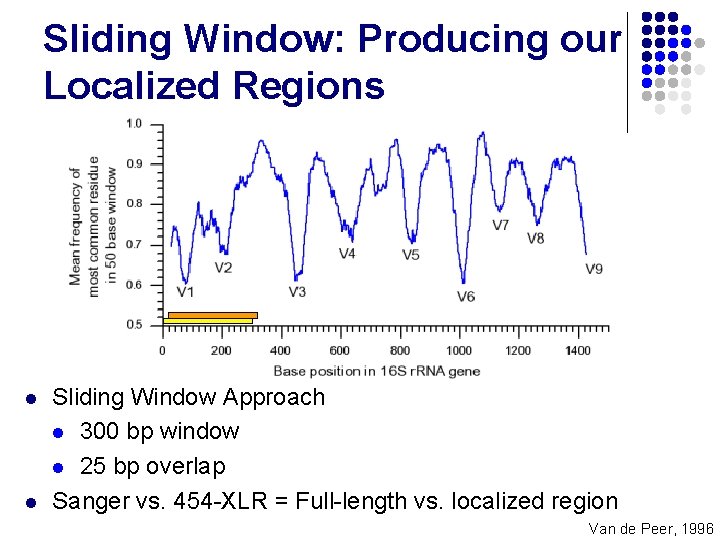
Sliding Window: Producing our Localized Regions l l Sliding Window Approach l 300 bp window l 25 bp overlap Sanger vs. 454 -XLR = Full-length vs. localized region Van de Peer, 1996

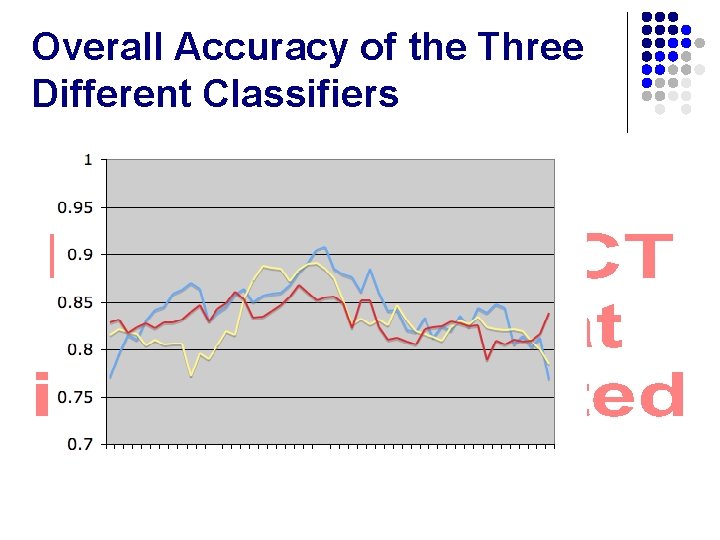
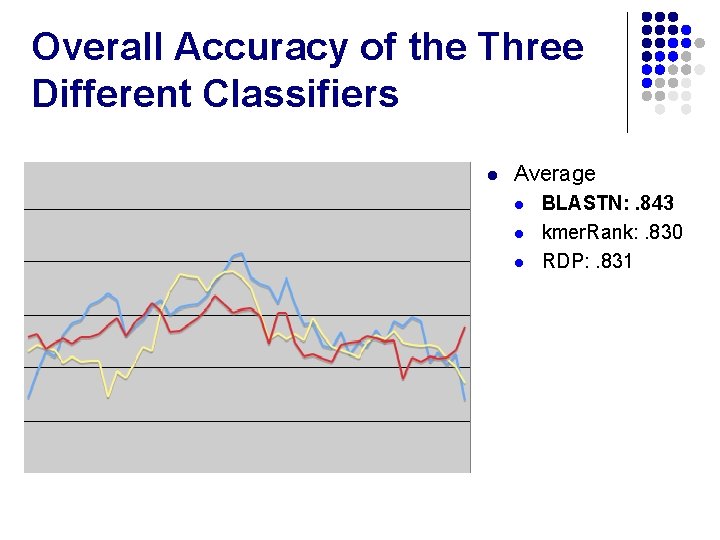
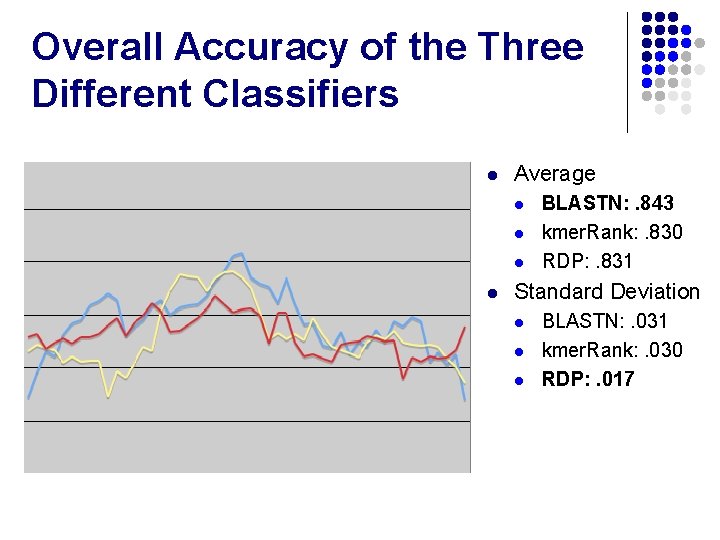
Overall Accuracy of the Three Different Classifiers

Overall Accuracy of the Three Different Classifiers l Average l l l BLASTN: . 843 kmer. Rank: . 830 RDP: . 831

Overall Accuracy of the Three Different Classifiers l Average l l BLASTN: . 843 kmer. Rank: . 830 RDP: . 831 Standard Deviation l l l BLASTN: . 031 kmer. Rank: . 030 RDP: . 017

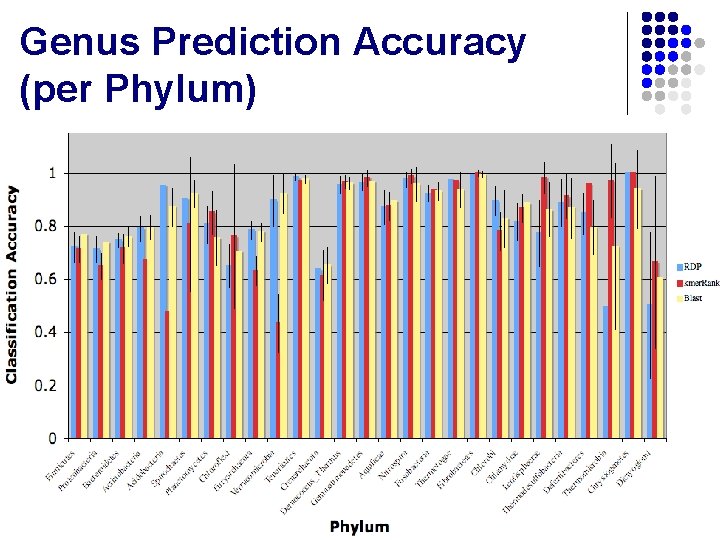
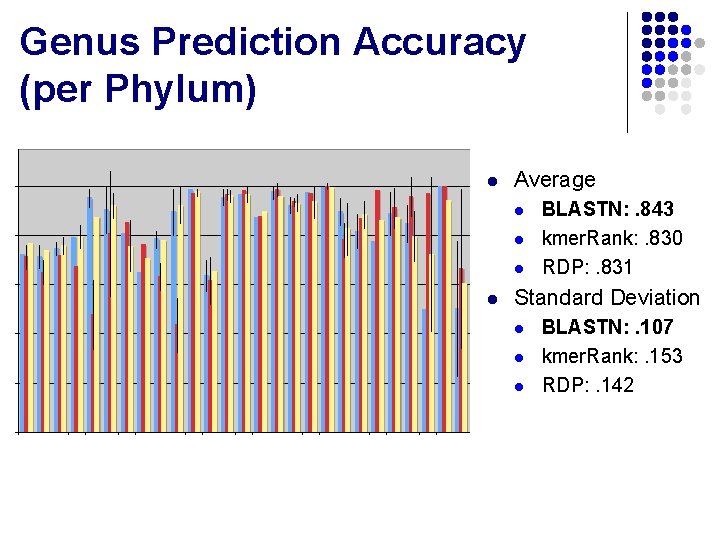
Genus Prediction Accuracy (per Phylum)

Genus Prediction Accuracy (per Phylum) l Average l l BLASTN: . 843 kmer. Rank: . 830 RDP: . 831 Standard Deviation l l l BLASTN: . 107 kmer. Rank: . 153 RDP: . 142

Finding the 16 S Region Providing the Most Reliable Prediction Accuracy

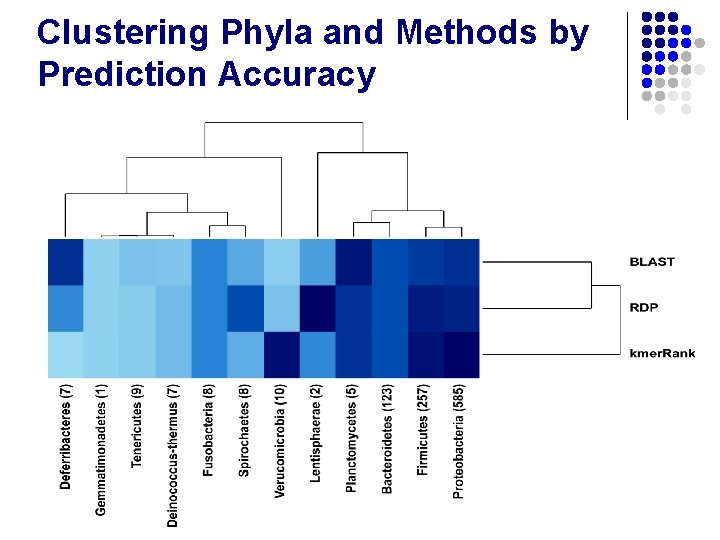
Clustering Phyla and Methods by Prediction Accuracy

Clustering Phyla and Methods by Prediction Accuracy l Best method is Phylum-dependent l Variation in accuracy impacted by depth of species coverage

Summary l l Central region of 16 S is the most accurate, on average Of the methods examined, BLAST is most accurate across all 16 S regions and all phyla, on average RDP-bayes is least variable across short sequence regions Best short sequence classification method is phylum-dependent

Acknowledgements l Genome Sequencing and Analysis Program l l l l Brian Haas Dirk Gevers Michael Feldgarden Doyle Ward Chad Nusbaum Bruce Birren Administration l l l Shawna Young Lucia Vielma Maura Silverstein
 Long and short
Long and short Microbiome nih
Microbiome nih Hình ảnh bộ gõ cơ thể búng tay
Hình ảnh bộ gõ cơ thể búng tay Bổ thể
Bổ thể Tỉ lệ cơ thể trẻ em
Tỉ lệ cơ thể trẻ em Gấu đi như thế nào
Gấu đi như thế nào Chụp phim tư thế worms-breton
Chụp phim tư thế worms-breton Chúa sống lại
Chúa sống lại Các môn thể thao bắt đầu bằng tiếng bóng
Các môn thể thao bắt đầu bằng tiếng bóng Thế nào là hệ số cao nhất
Thế nào là hệ số cao nhất Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Công thức tiính động năng
Công thức tiính động năng Trời xanh đây là của chúng ta thể thơ
Trời xanh đây là của chúng ta thể thơ Mật thư tọa độ 5x5
Mật thư tọa độ 5x5 Làm thế nào để 102-1=99
Làm thế nào để 102-1=99 độ dài liên kết
độ dài liên kết Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Thể thơ truyền thống
Thể thơ truyền thống Quá trình desamine hóa có thể tạo ra
Quá trình desamine hóa có thể tạo ra Một số thể thơ truyền thống
Một số thể thơ truyền thống Cái miệng nó xinh thế chỉ nói điều hay thôi
Cái miệng nó xinh thế chỉ nói điều hay thôi Vẽ hình chiếu vuông góc của vật thể sau
Vẽ hình chiếu vuông góc của vật thể sau