Vaccine Design Need for new vaccine technologies The









- Slides: 29

Vaccine Design

Need for new vaccine technologies • • The classical way of making vaccines have in many cases been tried for the pathogens for which no vaccines exist Need for new ways for making vaccines

Categories of Vaccines • Live vaccines • Are able to replicate in the host • Attenuated (weakened) so they do not cause disease • Subunit vaccines • Part of organism • Genetic Vaccines • Part of genes from organism

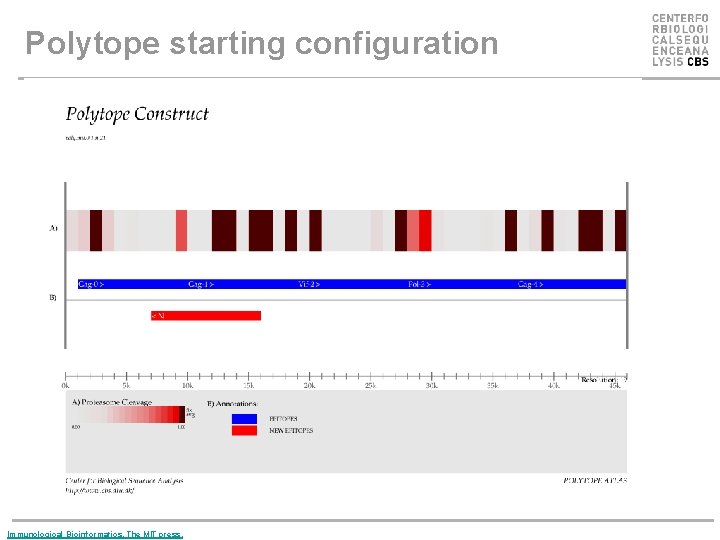
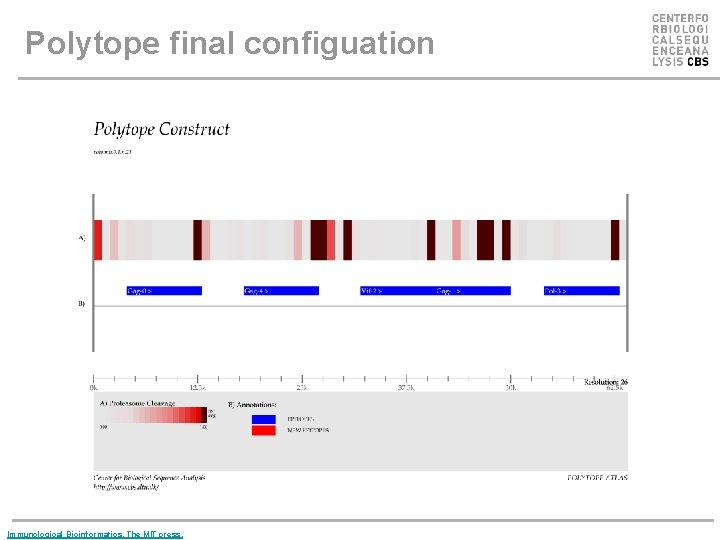
Polytope optimization • Successful immunization can be obtained only if the epitopes encoded by the polytope are correctly processed and presented. • Cleavage by the proteasome in the cytosol, translocation into the ER by the TAP complex, as well as binding to MHC class I should be taken into account in an integrative manner. • The design of a polytope can be done in an effective way by modifying the sequential order of the different epitopes, and by inserting specific amino acids that will favor optimal cleavage and transport by the TAP complex, as linkers between the epitopes.

Polytope starting configuration Immunological Bioinformatics, The MIT press.

Polytope optimization Algorithm • Optimization of of four measures: 1. The number of poor C-terminal cleavage sites of epitopes (predicted cleavage < 0. 9) 2. The number of internal cleavage sites (within epitope cleavages with a prediction larger than the predicted C-terminal cleavage) 3. The number of new epitopes (number of processed and presented epitopes in the fusing regions spanning the epitopes) 4. The length of the linker region inserted between epitopes. • The optimization seeks to minimize the above four terms by use of Monte Carlo Metropolis simulations [Metropolis et al. , 1953]

Polytope final configuation Immunological Bioinformatics, The MIT press.

Prediction of antigens • Protective antigens • Functional definition (phenotype) • Which antigens will be protective (genotype)? • They must be recognized by the immune system • Predict epitopes (include processing) • CTL (MHC class I) • http: //www. cbs. dtu. dk/services/Net. CTL/ • Helper (MHC class II) • http: //mail 1. imtech. res. in/raghava/hlapred/index. html • Antibody • http: //www. cbs. dtu. dk/services/Bepi. Pred/ • http: //www. cbs. dtu. dk/services/Disco. Tope/ More Links: http: //www. cbs. dtu. dk/researchgroups/immunology/webreview. html

Function and conservation • Some of the epitopes must exist in the wild type • Conservation • http: //www. ncbi. nlm. nih. gov/BLAST/ • Function • When is it expressed? • Where is it trafficked to? • Secretome. P Non-classical and leaderless secretion of eukaryotic proteins. Signal. P Signal peptide and cleavage sites in gram+, gramand eukaryotic amino acid sequences. Target. P Subcellular location of proteins: mitochondrial, chloroplastic, secretory pathway, or other. • Expression level?

Selection of antigens • Epitopes • Polytope • Proteins • Helper epitopes • Does it contain any • Can they be added • Hide epitopes • Immunodominant and variable ones

Examples of antigen selections

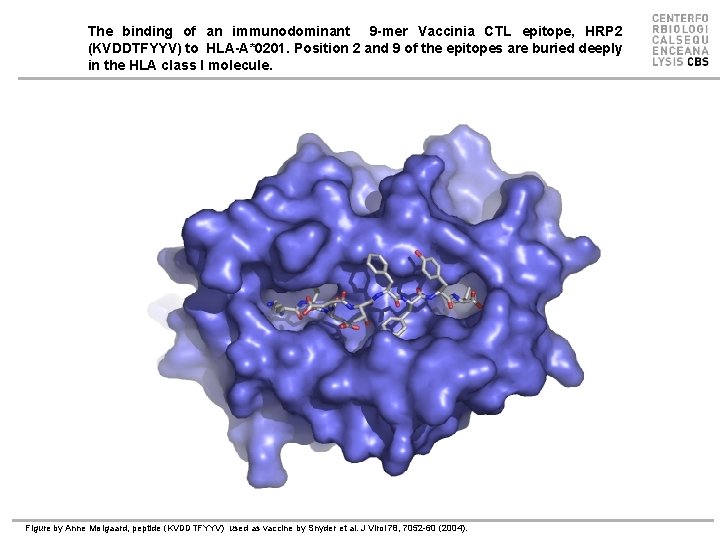
The binding of an immunodominant 9 -mer Vaccinia CTL epitope, HRP 2 (KVDDTFYYV) to HLA-A*0201. Position 2 and 9 of the epitopes are buried deeply in the HLA class I molecule. Figure by Anne Mølgaard, peptide (KVDDTFYYV) used as vaccine by Snyder et al. J Virol 78, 7052 -60 (2004).


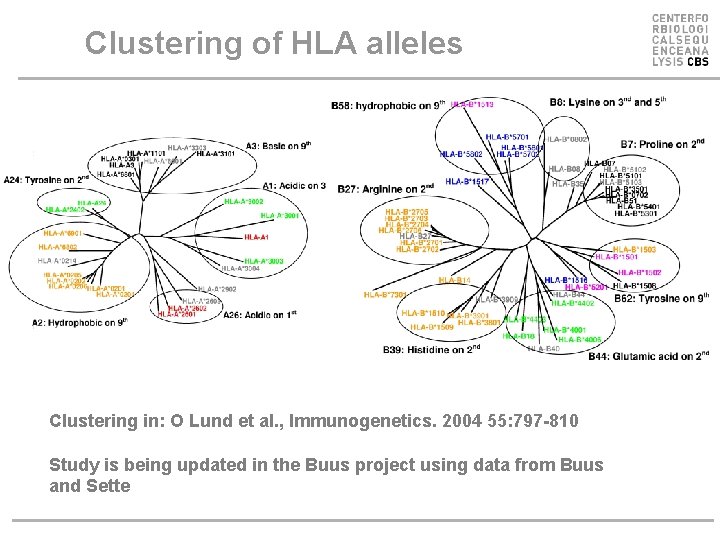
Clustering of HLA alleles Clustering in: O Lund et al. , Immunogenetics. 2004 55: 797 -810 Study is being updated in the Buus project using data from Buus and Sette

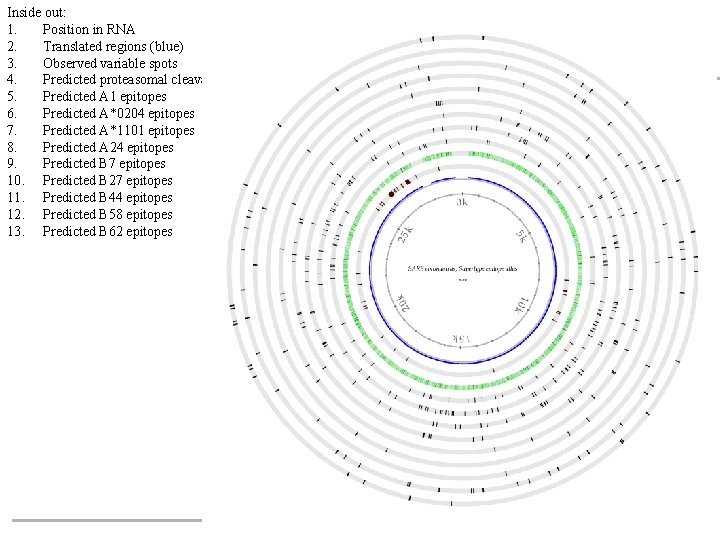
Inside out: 1. Position in RNA 2. Translated regions (blue) 3. Observed variable spots 4. Predicted proteasomal cleavage 5. Predicted A 1 epitopes 6. Predicted A*0204 epitopes 7. Predicted A*1101 epitopes 8. Predicted A 24 epitopes 9. Predicted B 7 epitopes 10. Predicted B 27 epitopes 11. Predicted B 44 epitopes 12. Predicted B 58 epitopes 13. Predicted B 62 epitopes

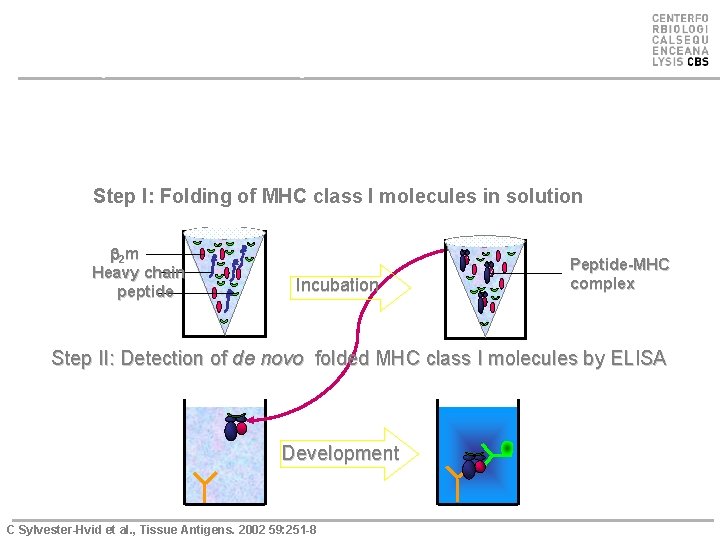
Strategy for the quantitative ELISA assay C. Sylvester-Hvid, et al. , Tissue antigens, 2002: 59: 251 Step I: Folding of MHC class I molecules in solution b 2 m Heavy chain peptide Incubation Peptide-MHC complex Step II: Detection of de novo folded MHC class I molecules by ELISA Development C Sylvester-Hvid et al. , Tissue Antigens. 2002 59: 251 -8

SARS project l. We scanned HLA supertypes and identified almost 100 potential vaccine candidates. l. These should be further validated in SARS survivors and may be used for vaccine formulation. l. Prediction method available: www. cbs. dtu. dk/services/Net. MHC/ C Sylvester-Hvid et al. , Tissue Antigens. 2004 63: 395 -400

NIH project Develop improved methods to predict cytotoxic T cell (CTL) epitopes Scan 15 different pathogens from the NIAID A-C list agents of bioterrorism Test if cytotoxic T cells from preselected immune blood donors can react to the selected peptides for 3 selected pathogens: Influenza, Smallpox vaccine and tuberculosis vaccine (BCG)

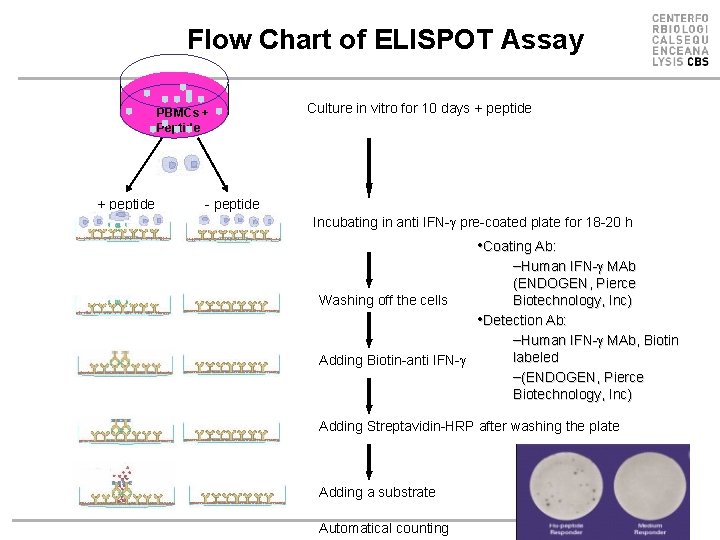
Flow Chart of ELISPOT Assay PBMCs + Peptide + peptide Culture in vitro for 10 days + peptide - peptide Incubating in anti IFN- pre-coated plate for 18 -20 h • Coating Ab: –Human IFN- MAb (ENDOGEN, Pierce Biotechnology, Inc) Washing off the cells • Detection Ab: –Human IFN- MAb, Biotin labeled Adding Biotin-anti IFN- –(ENDOGEN, Pierce Biotechnology, Inc) Adding Streptavidin-HRP after washing the plate Adding a substrate Automatical counting

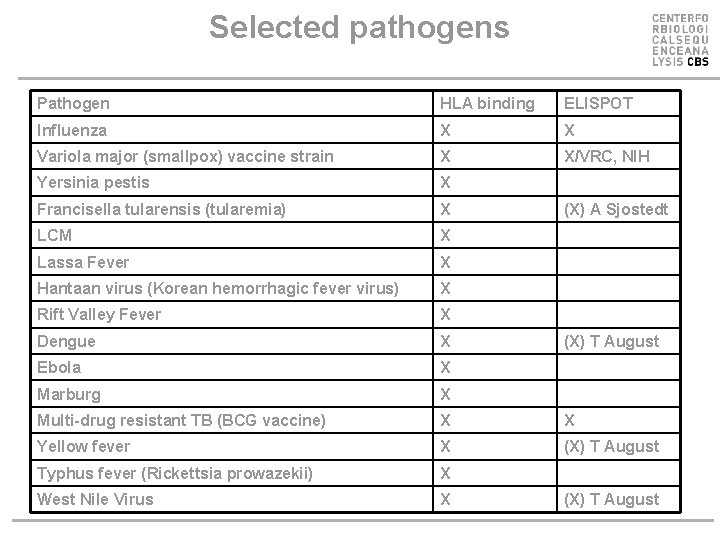
Selected pathogens Pathogen HLA binding ELISPOT Influenza X X Variola major (smallpox) vaccine strain X X/VRC, NIH Yersinia pestis X Francisella tularensis (tularemia) X LCM X Lassa Fever X Hantaan virus (Korean hemorrhagic fever virus) X Rift Valley Fever X Dengue X Ebola X Marburg X Multi-drug resistant TB (BCG vaccine) X X Yellow fever X (X) T August Typhus fever (Rickettsia prowazekii) X West Nile Virus X (X) A Sjostedt (X) T August

Prediction of Class II epitopes

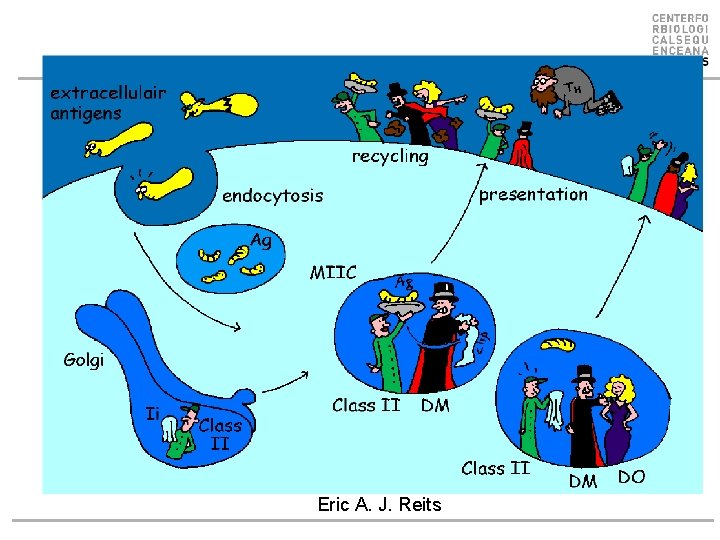
Eric A. J. Reits

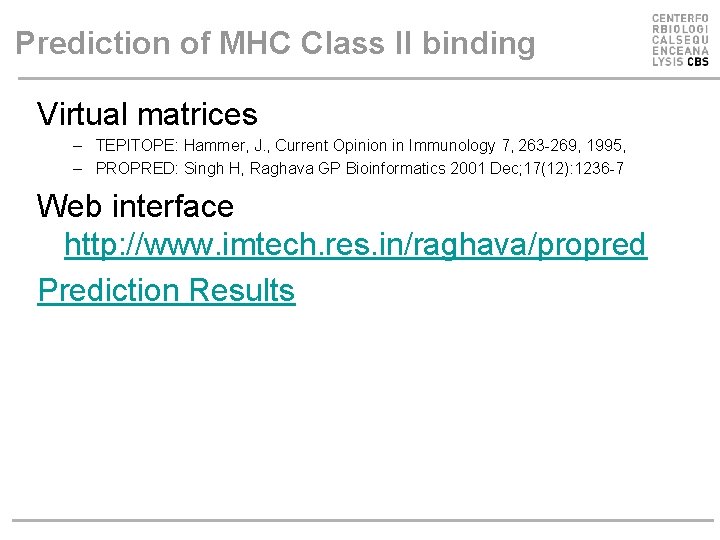
Prediction of MHC Class II binding Virtual matrices – TEPITOPE: Hammer, J. , Current Opinion in Immunology 7, 263 -269, 1995, – PROPRED: Singh H, Raghava GP Bioinformatics 2001 Dec; 17(12): 1236 -7 Web interface http: //www. imtech. res. in/raghava/propred Prediction Results

Prediction of Antibody epitopes Linear – Hydrophilicity scales (average in ~7 window) • Hoop and Woods (1981) • Kyte and Doolittle (1982) • Parker et al. (1986) – Other scales & combinations • Pellequer and van Regenmortel • Alix – New improved method (Pontoppidan et al. in preparation) • http: //www. cbs. dtu. dk/services/Bepi. Pred/ Discontinuous – Protrusion (Novotny, Thornton, 1986)



Prediction of proteins structure • Homology modeling • Secondary structure prediction


Acknowledgements Immunological Bioinformatics group, CBS, Technical University of Denmark (www. cbs. dtu. dk) Morten Nielsen HLA binding Claus Lundegaard Data bases, HLA binding Anne Mølgaard MHC structure Mette Voldby Larsen Phd student - CTL prediction Pernille Haste Andersen Ph. D student – Structure Sune Frankild Ph. D student - Databases Jens Pontoppidan Linear B cell epitopes Thomas Blicher MHC structure Sheila Tang Pox/TB Thomas Rask Evolution Nicolas Rapin/Ilka Hoff Simulation of the immune system Collaborators IMMI, University of Copenhagen Søren Buus MHC binding Mogens H Claesson Elispot Assay La Jolla Institute of Allergy and Infectious Diseases Allesandro Sette Epitope database Bjoern Peters Leiden University Medical Center Tom Ottenhoff Tuberculosis Michel Klein Fatima Kazi Ganymed Ugur Sahin Genetic library University of Tubingen Stefan Stevanovic MHC ligands INSERM Peter van Endert Tap binding University of Mainz Hansjörg Schild Proteasome Schafer-Nielsen Claus Schafer-Nielsen Peptide synthesis University of Utrecht Can Kesmir Bioinformatics
 Immigration and urbanization new technologies lesson 4
Immigration and urbanization new technologies lesson 4 New disruptive technologies 2021
New disruptive technologies 2021 New disruptive technologies
New disruptive technologies Disjunctive questions
Disjunctive questions Formuö
Formuö Typiska novell drag
Typiska novell drag Nationell inriktning för artificiell intelligens
Nationell inriktning för artificiell intelligens Vad står k.r.å.k.a.n för
Vad står k.r.å.k.a.n för Varför kallas perioden 1918-1939 för mellankrigstiden?
Varför kallas perioden 1918-1939 för mellankrigstiden? En lathund för arbete med kontinuitetshantering
En lathund för arbete med kontinuitetshantering Kassaregister ideell förening
Kassaregister ideell förening Personlig tidbok fylla i
Personlig tidbok fylla i A gastrica
A gastrica Vad är densitet
Vad är densitet Datorkunskap för nybörjare
Datorkunskap för nybörjare Stig kerman
Stig kerman Tes debattartikel
Tes debattartikel Magnetsjukhus
Magnetsjukhus Nyckelkompetenser för livslångt lärande
Nyckelkompetenser för livslångt lärande Påbyggnader för flakfordon
Påbyggnader för flakfordon Arkimedes princip formel
Arkimedes princip formel Svenskt ramverk för digital samverkan
Svenskt ramverk för digital samverkan Jag har gått inunder stjärnor text
Jag har gått inunder stjärnor text Presentera för publik crossboss
Presentera för publik crossboss Argument för teckenspråk som minoritetsspråk
Argument för teckenspråk som minoritetsspråk Vem räknas som jude
Vem räknas som jude Treserva lathund
Treserva lathund Epiteltyper
Epiteltyper Claes martinsson
Claes martinsson Cks
Cks