Regulation of Gene Expression Prof Sezgin Gne Department




- Slides: 39

Regulation of Gene Expression Prof Sezgin Güneş Department of Medical Biology

Contents § Gene Expression § Regulation of Gene Expression in Prokaryotes § Regulation of Gene Expression in Eukaryotes 2

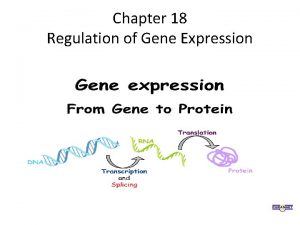
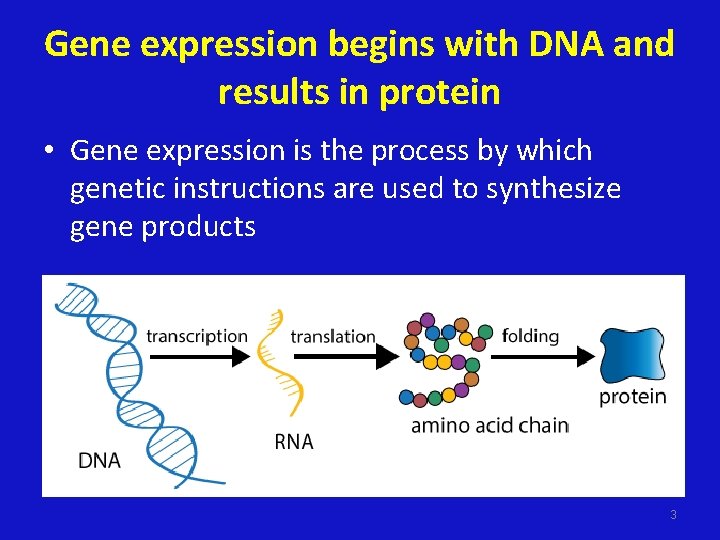
Gene expression begins with DNA and results in protein • Gene expression is the process by which genetic instructions are used to synthesize gene products 3

Gene Expression § The process by which a gene's information is converted into the structures and functions of a cell by a process of producing a biologically functional molecule of either protein or RNA is made § Gene expression is assumed to be controlled at various points in the sequence leading to protein synthesis 4

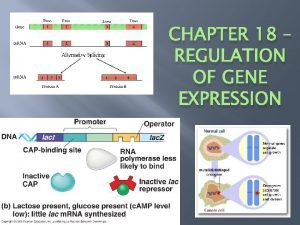
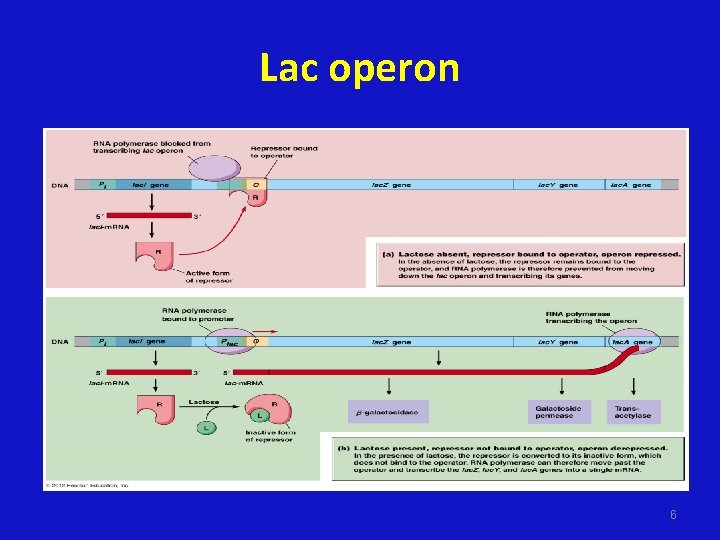
Lac operon 5

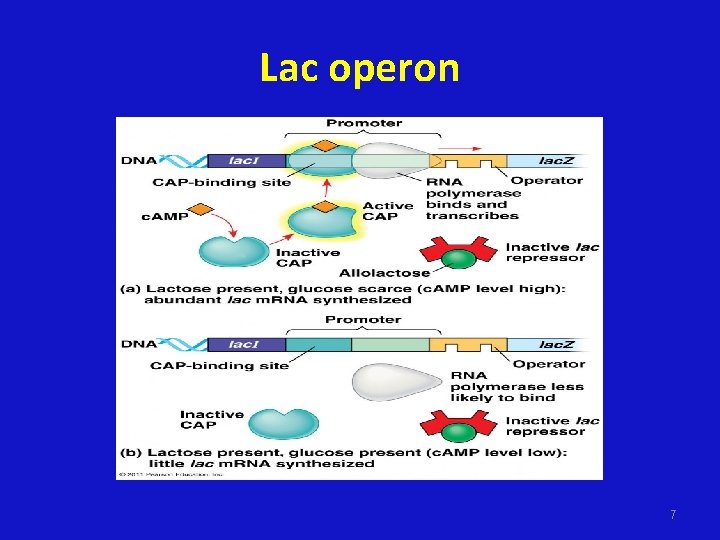
Lac operon 6

Lac operon 7

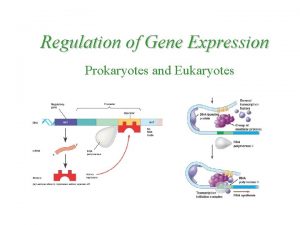
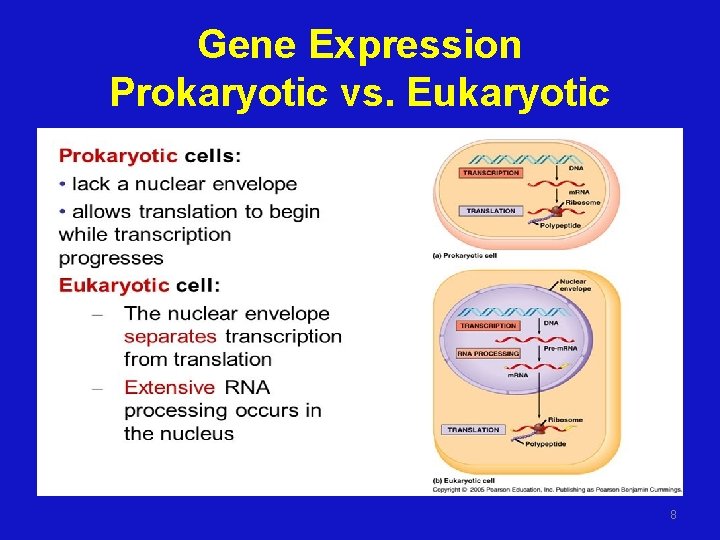
Gene Expression Prokaryotic vs. Eukaryotic 8

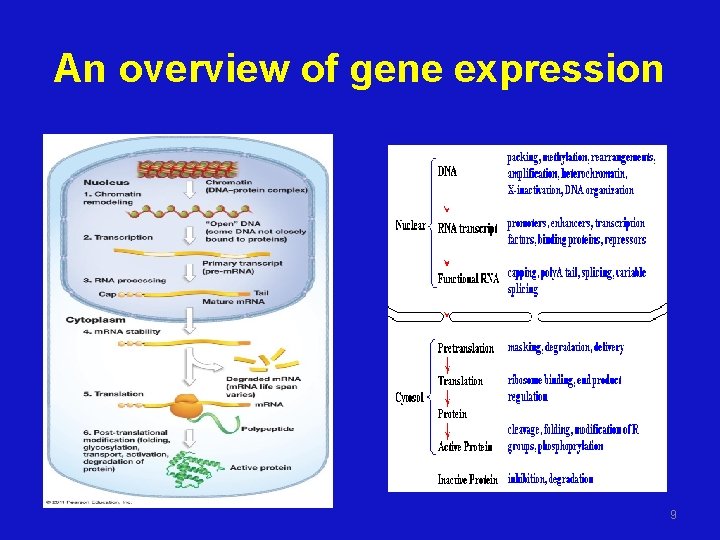
An overview of gene expression 9

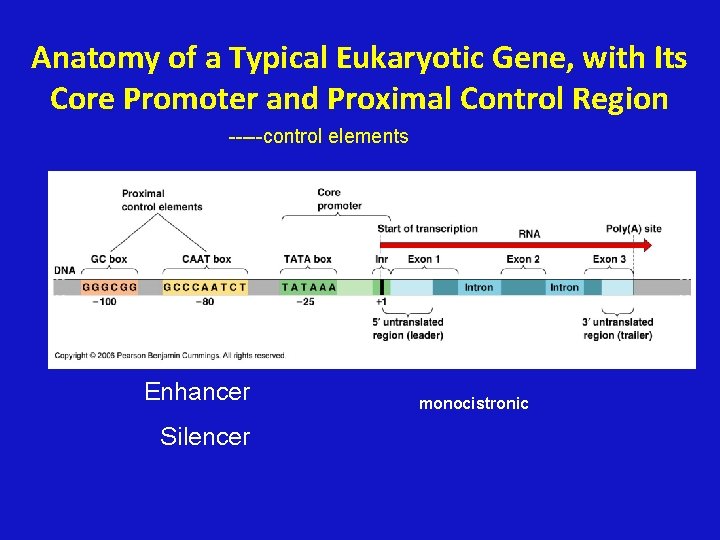
Anatomy of a Typical Eukaryotic Gene, with Its Core Promoter and Proximal Control Region -----control elements Enhancer Silencer monocistronic

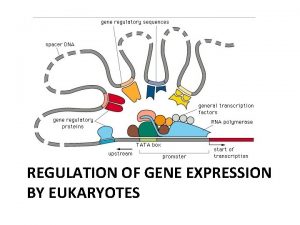
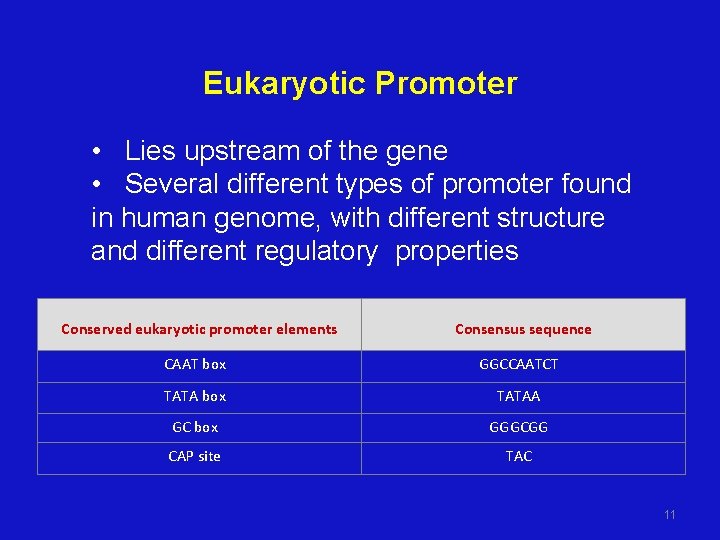
Eukaryotic Promoter • Lies upstream of the gene • Several different types of promoter found in human genome, with different structure and different regulatory properties Conserved eukaryotic promoter elements Consensus sequence CAAT box GGCCAATCT TATA box TATAA GC box GGGCGG CAP site TAC 11

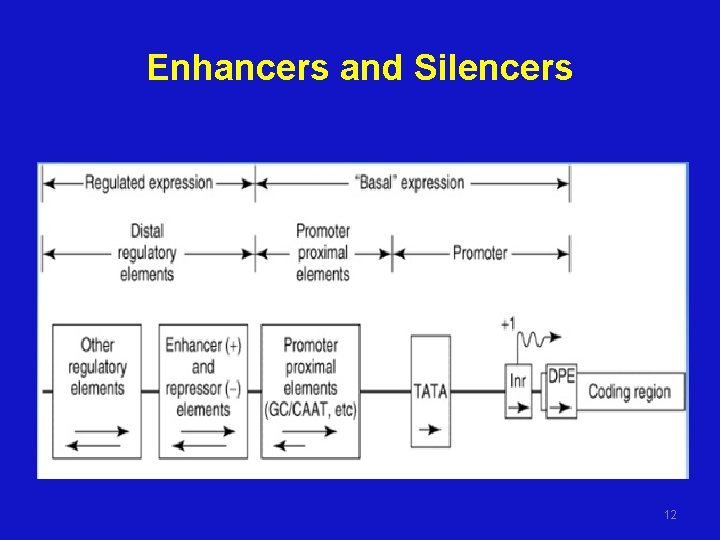
Enhancers and Silencers 12

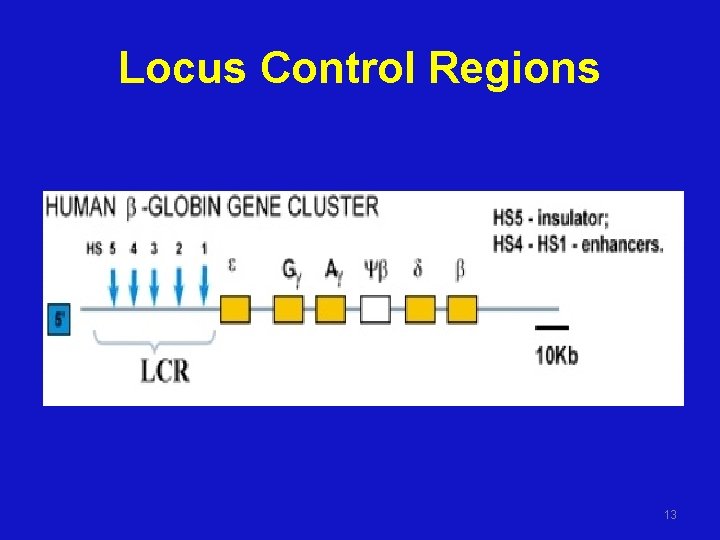
Locus Control Regions 13

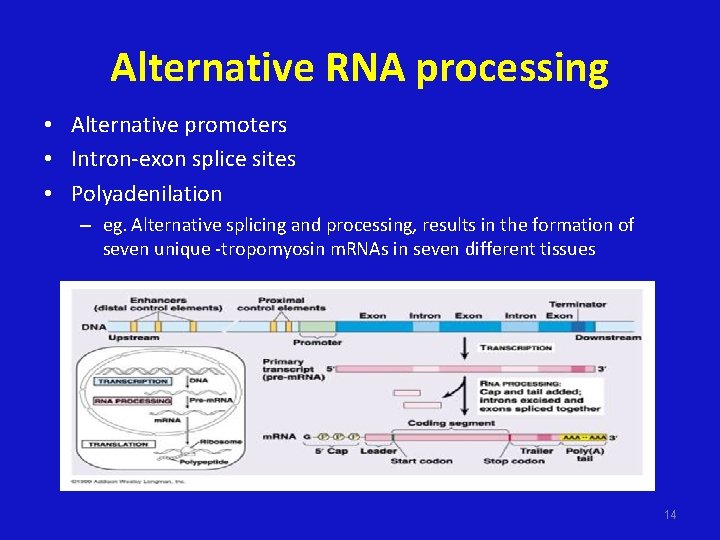
Alternative RNA processing • Alternative promoters • Intron-exon splice sites • Polyadenilation – eg. Alternative splicing and processing, results in the formation of seven unique -tropomyosin m. RNAs in seven different tissues 14

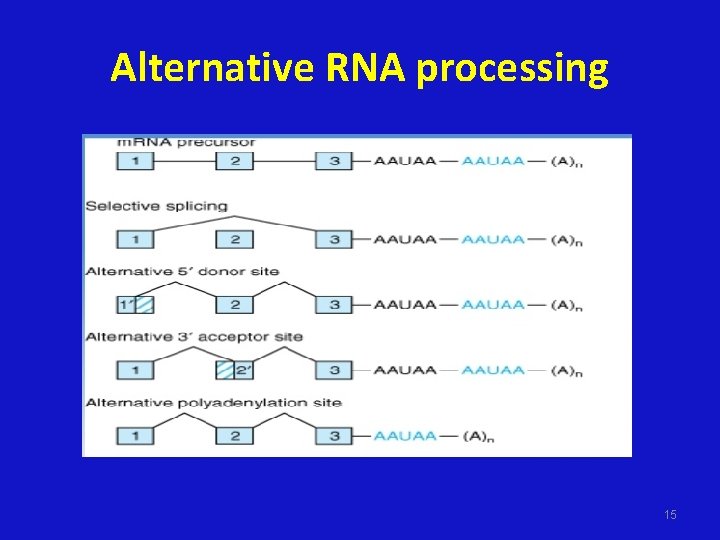
Alternative RNA processing 15

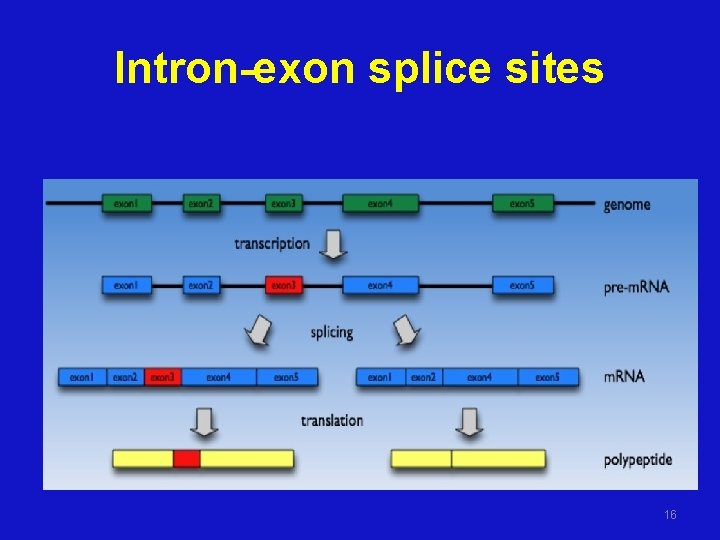
Intron-exon splice sites 16

m. RNA stability • Turn-over rate very short (~ 30 min) • The stability of the m. RNA can be influenced by hormones and certain other effectors • The ends of m. RNA molecules are involved in m. RNA stability – The 5' cap structure in eukaryotic m. RNA prevents attack by 5' exonucleases – the poly(A) tail prohibits the action of 3‘ exonucleases 17

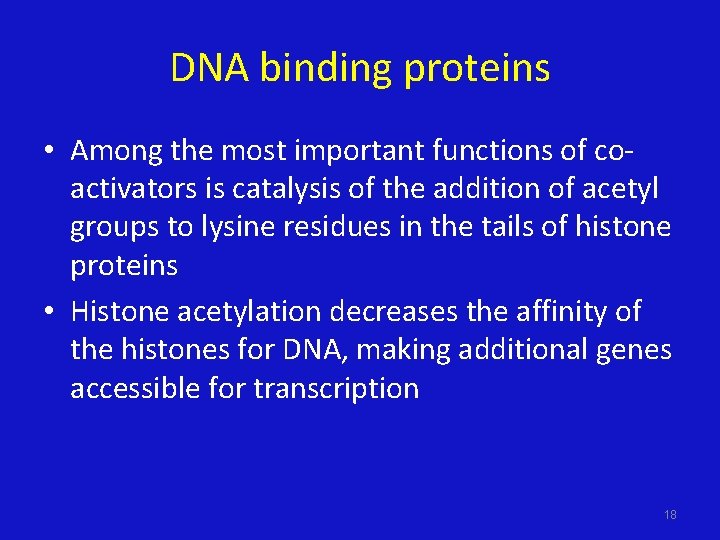
DNA binding proteins • Among the most important functions of coactivators is catalysis of the addition of acetyl groups to lysine residues in the tails of histone proteins • Histone acetylation decreases the affinity of the histones for DNA, making additional genes accessible for transcription 18

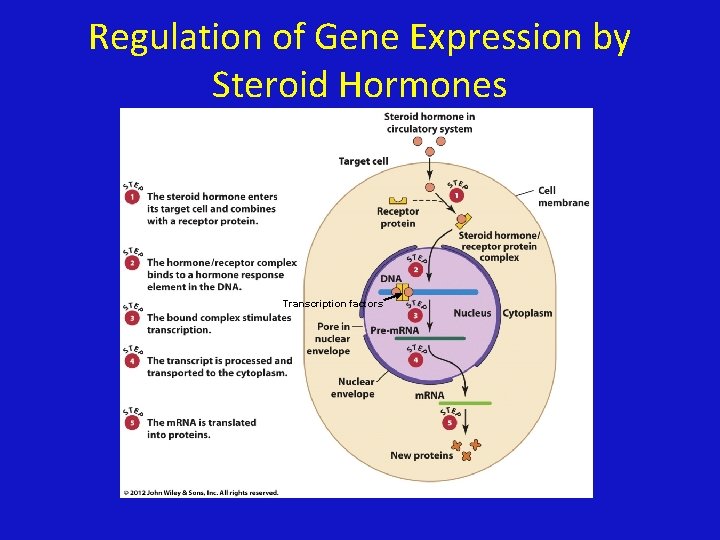
Regulation of Gene Expression by Steroid Hormones Transcription factors

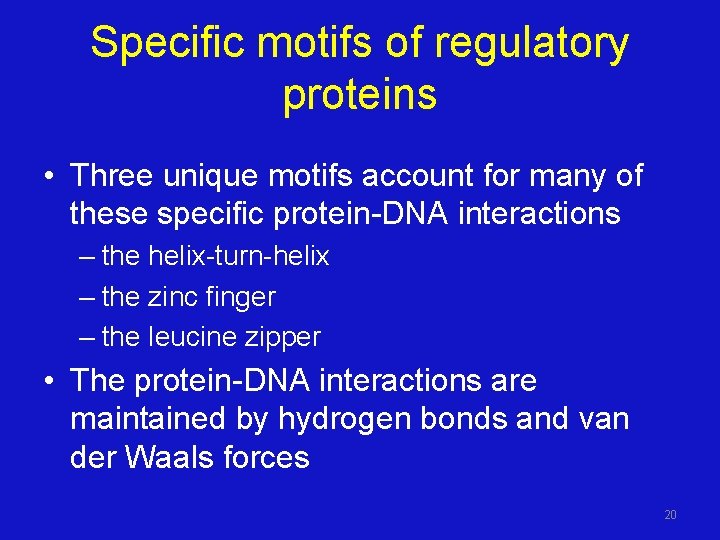
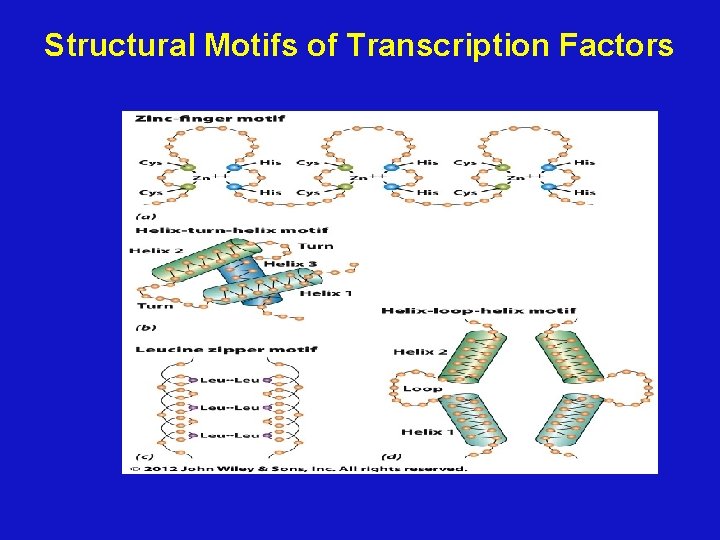
Specific motifs of regulatory proteins • Three unique motifs account for many of these specific protein-DNA interactions – the helix-turn-helix – the zinc finger – the leucine zipper • The protein-DNA interactions are maintained by hydrogen bonds and van der Waals forces 20

Structural Motifs of Transcription Factors

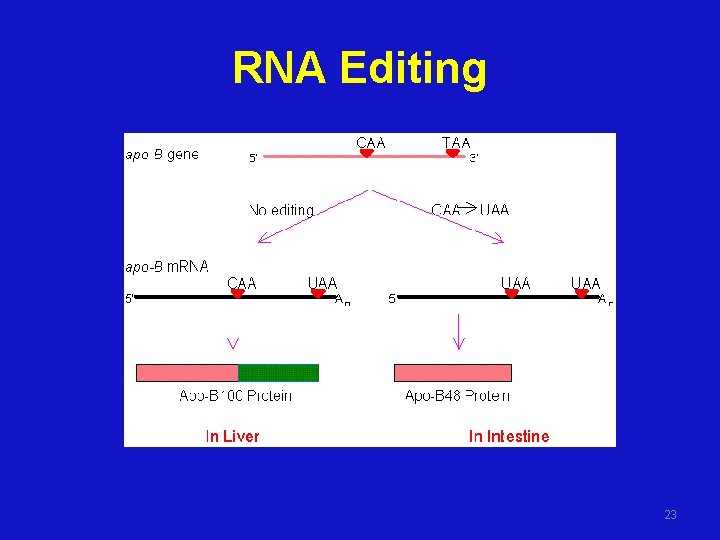
RNA Editing • Enzyme- catalyzed deamination of a specific cytidine residue in the m. RNA of apolipoprotein B-100 changes a codon for glutamine (CAA) to a stop codon (UAA) • Apolipoprotein B-48, a truncated version of the protein lacking the LDL receptorbinding domain, is generated by this posttranscriptional change in the m. RNA sequence 22

RNA Editing 23

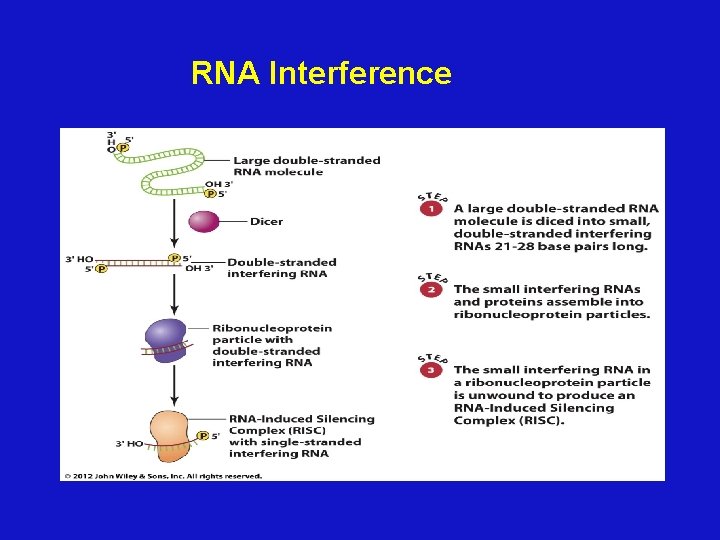
RNA Interference • Small, noncoding RNAs base pair with target sequences in m. RNA • The small RNAs interfere with expression of the target m. RNAs

RNA Interference


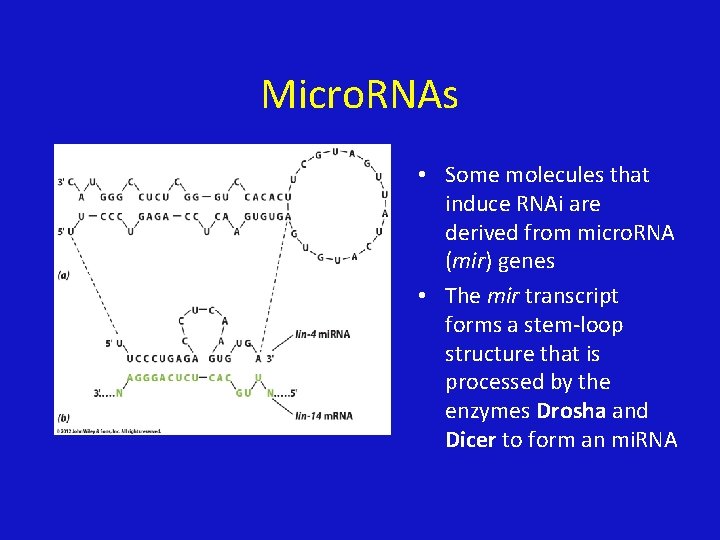
Micro. RNAs • Some molecules that induce RNAi are derived from micro. RNA (mir) genes • The mir transcript forms a stem-loop structure that is processed by the enzymes Drosha and Dicer to form an mi. RNA

Sources of si. RNA and mi. RNA • mi. RNAs are derived from endogenous transcripts of the mir genes • Long double-stranded RNA that is transfected or injected into a cell can be processed to form an si. RNA. This can be used experimentally to knock down expression of a gene.

Translation § Translation is the process by which ribosomes read the genetic message in the m. RNA and produce a protein product according to the message's instruction 29

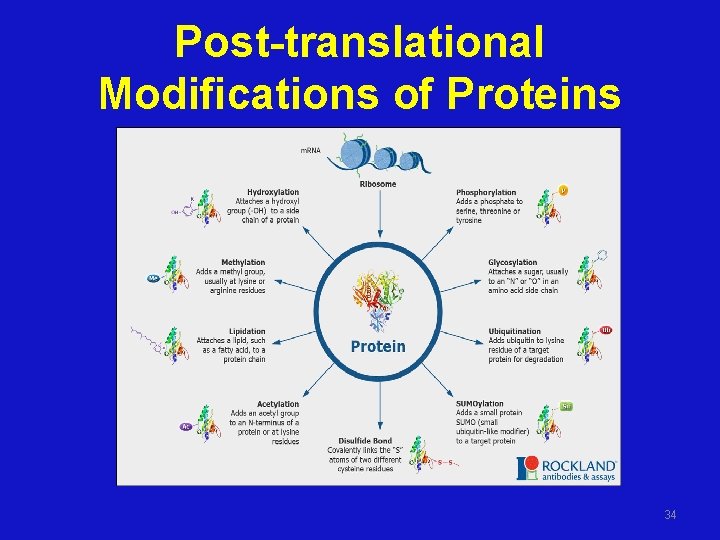
Post-translational Modifications of Proteins • Alteration of proteins after translation • Give functional activity – Peptide cleavage – Covalent modifications • Destination of proteins – Signal peptide – Nuclear targetting signals 30

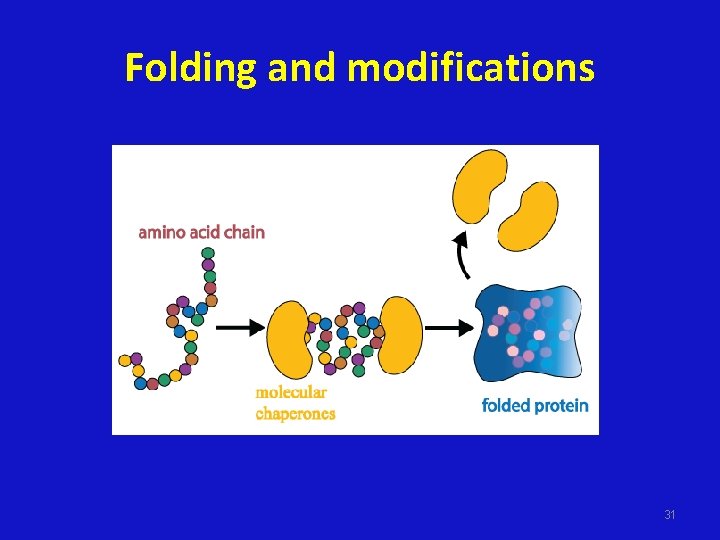
Folding and modifications 31

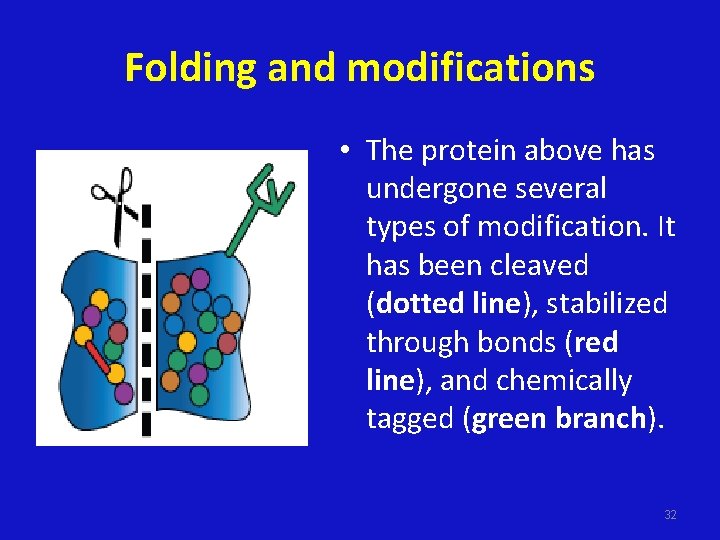
Folding and modifications • The protein above has undergone several types of modification. It has been cleaved (dotted line), stabilized through bonds (red line), and chemically tagged (green branch). 32

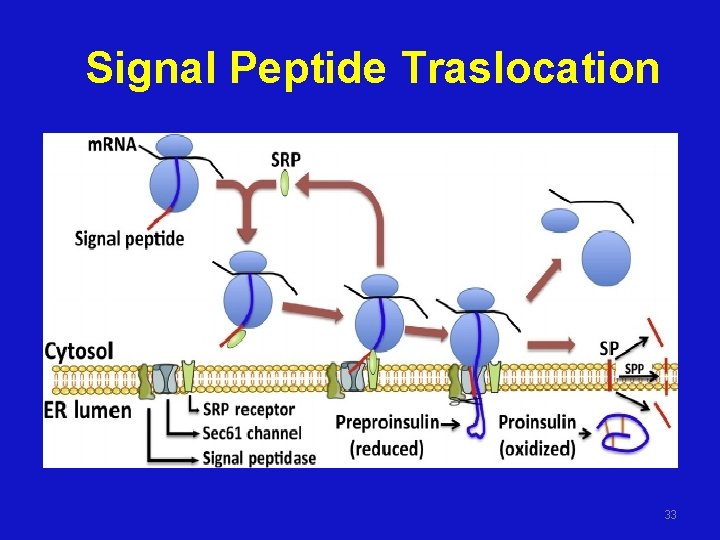
Signal Peptide Traslocation 33

Post-translational Modifications of Proteins 34

Sumary • The genetic constitutions of nearly all metazoan somatic cells are identical. • Tissue or cell specificity is dictated by differences in gene expression of this complement of genes. • Alterations in gene expression allow a cell to adapt to environmental changes. • Gene expression can be controlled at multiple levels by chromatin modifications , changes in transcription, RNA processing, localization, and stability or utilization. • Gene amplification and rearrangements also influence gene expression. 35

Eukaryotic Gene Expression § Essentially all humans' genes contain introns. A § § § notable exception is the histone genes which are intronless Eukaryote genes are not grouped in operons. Each eukaryote gene is transcribed separately, with separate transcriptional controls on each gene Eukaryotic m. RNA is modified through RNA splicing Eukaryotic m. RNA is generally monogenic (monocistronic); code for only one polypeptide. 36

Eukaryotic Gene Expression § Eukaryotic m. RNA contain no Shine-Dalgarno § § § sequence to show the ribosomes where to start translating. Instead, most eukaryotic m. RNA have caps at their 5` end which directs initiation factors to bind and begin searching for an initiation codon. Eukaryotes have a separate RNA polymerase for each type of RNA. In eukaryotes, polysomes are found in the cytoplasm. Eukaryotic protein synthesis initiation begins with methionine not N formyl- methionine. 37

Prokaryotic vs. Eukaryotic § Prokaryote genes are grouped in operons § Prokaryotes have one type of RNA polymerase for all types of RNA § m. RNA is not modified § The existence of introns in prokaryotes is extremely rare 38

Prokaryotic vs. Eukaryotic §To initiate transcription in bacteria, sigma factors bind to RNA polymerases/ sigma factors complex can then bind to promoter about 40 deoxyribonucleotide bases prior to the coding region of the gene §In prokaryotes, the newly synthesized m. RNA is polycistronic (polygenic) (code for more than one polypeptide chain) §In prokaryotes, transcription of a gene and translation of the resulting m. RNA occur simultaneously. So many polysomes are found associated with an active gene 39
 Prof. dr. sezgin alsan
Prof. dr. sezgin alsan Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Regulation of gene expression
Regulation of gene expression Regulation of gene expression in bacteria
Regulation of gene expression in bacteria Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Chapter 18: regulation of gene expression
Chapter 18: regulation of gene expression Chapter 17 from gene to protein
Chapter 17 from gene to protein Gne farm equipment
Gne farm equipment Gne enerji eski tip
Gne enerji eski tip Section 4 gene regulation and mutation
Section 4 gene regulation and mutation Section 12-5 gene regulation answer key
Section 12-5 gene regulation answer key Section 4 gene regulation and mutations
Section 4 gene regulation and mutations Lac operon positive or negative control
Lac operon positive or negative control Gene regulation
Gene regulation What is gene regulation
What is gene regulation 12-5 gene regulation
12-5 gene regulation Gene regulation
Gene regulation Prokaryotes vs eukaryotes gene regulation
Prokaryotes vs eukaryotes gene regulation Differential gene regulation
Differential gene regulation Lojik
Lojik Gene by gene test results
Gene by gene test results Gene expression
Gene expression At dna
At dna Lac operon in prokaryotes
Lac operon in prokaryotes Real time pcr primer efficiency test
Real time pcr primer efficiency test Lyonization of gene expression
Lyonization of gene expression Prokaryotic
Prokaryotic Gene expression
Gene expression Gene expression omnibus tutorial
Gene expression omnibus tutorial What is the first step in gene expression
What is the first step in gene expression Gene expression
Gene expression Gene expression
Gene expression Genetic effects on gene expression across human tissues
Genetic effects on gene expression across human tissues Michigan licensing and regulatory affairs
Michigan licensing and regulatory affairs Ponzi scheme vs pyramid scheme
Ponzi scheme vs pyramid scheme Wa department of water and environmental regulation
Wa department of water and environmental regulation Quadratic equation examples
Quadratic equation examples Hong kong private placement regulation
Hong kong private placement regulation Rules for writing formal letters
Rules for writing formal letters