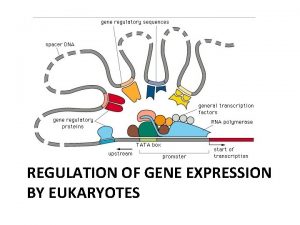
Gene Regulation Regulation of gene expression refers to







- Slides: 21

Gene Regulation

Regulation of gene expression • refers to the cellular control of the amount and timing of changes to the appearance of the functional product of a gene. Basically, anything that can regulate for example • – – Chemical and structural modification of DNA or chromatin Transcription Post-transcriptional modification m. RNA degradation

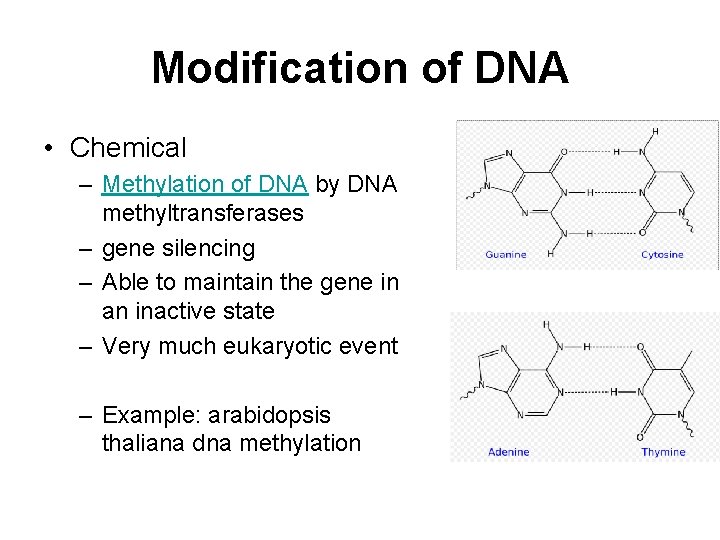
Modification of DNA • Chemical – Methylation of DNA by DNA methyltransferases – gene silencing – Able to maintain the gene in an inactive state – Very much eukaryotic event – Example: arabidopsis thaliana dna methylation

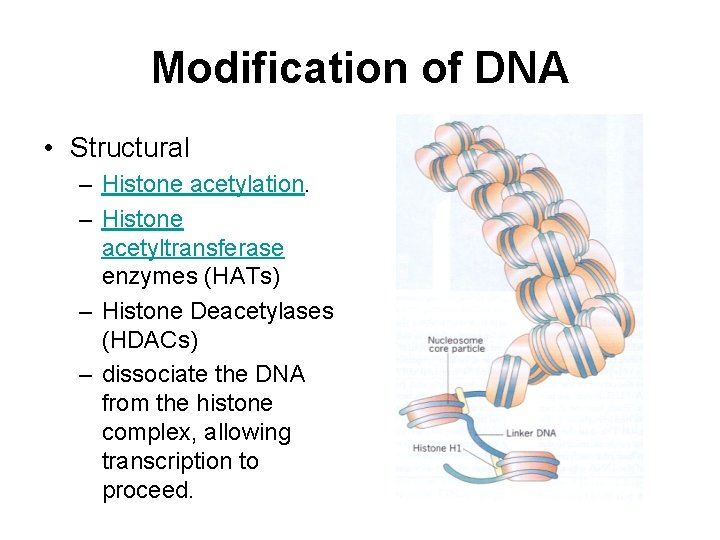
Modification of DNA • Structural – Histone acetylation. – Histone acetyltransferase enzymes (HATs) – Histone Deacetylases (HDACs) – dissociate the DNA from the histone complex, allowing transcription to proceed.


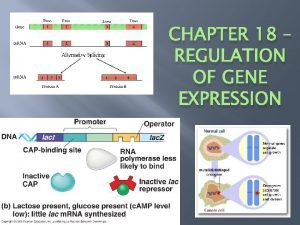
Regulation of transcription • Regulation of transcription controls when transcription occurs and how much RNA is created. Transcription of a gene by RNA polymerase can be regulated by at least five mechanisms: 1. Specificity factors 2. Repressors 3. General transcription factors 4. Activators 5. Enhancers

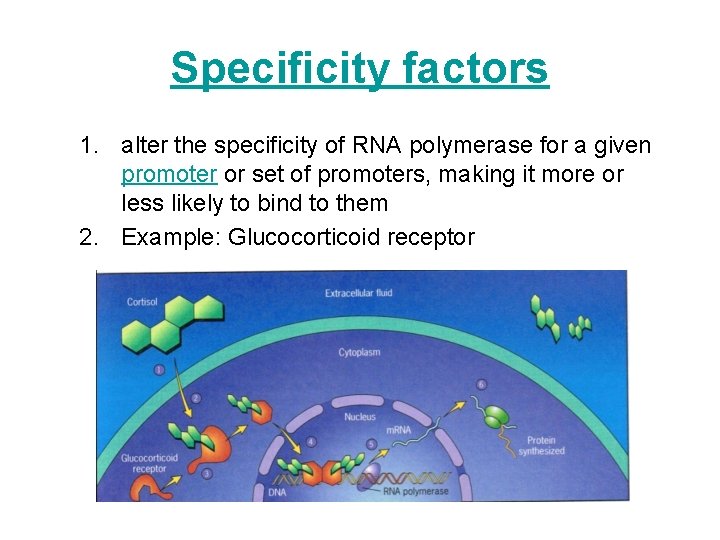
Specificity factors 1. alter the specificity of RNA polymerase for a given promoter or set of promoters, making it more or less likely to bind to them 2. Example: Glucocorticoid receptor

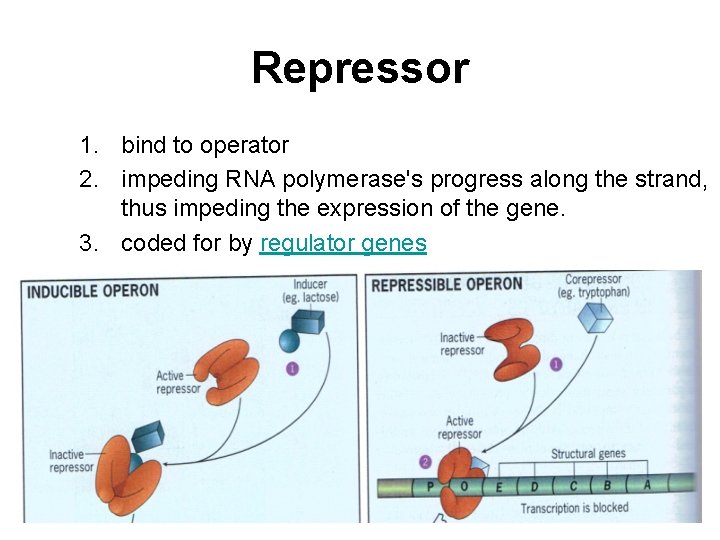
Repressor 1. bind to operator 2. impeding RNA polymerase's progress along the strand, thus impeding the expression of the gene. 3. coded for by regulator genes

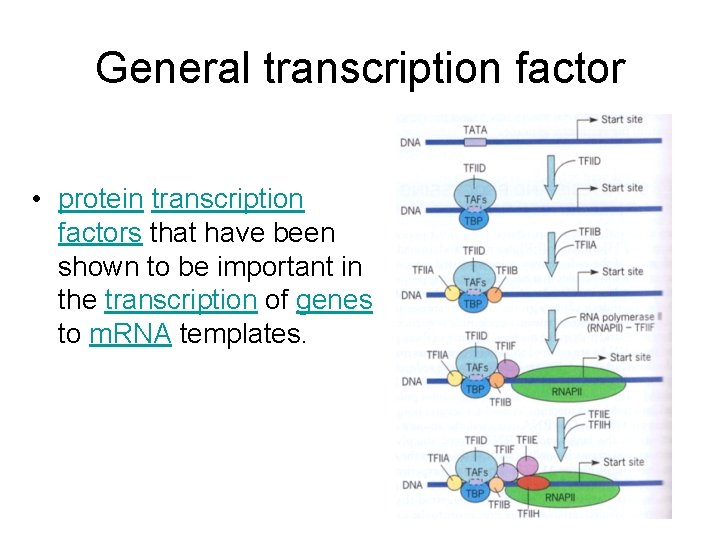
General transcription factor • protein transcription factors that have been shown to be important in the transcription of genes to m. RNA templates.

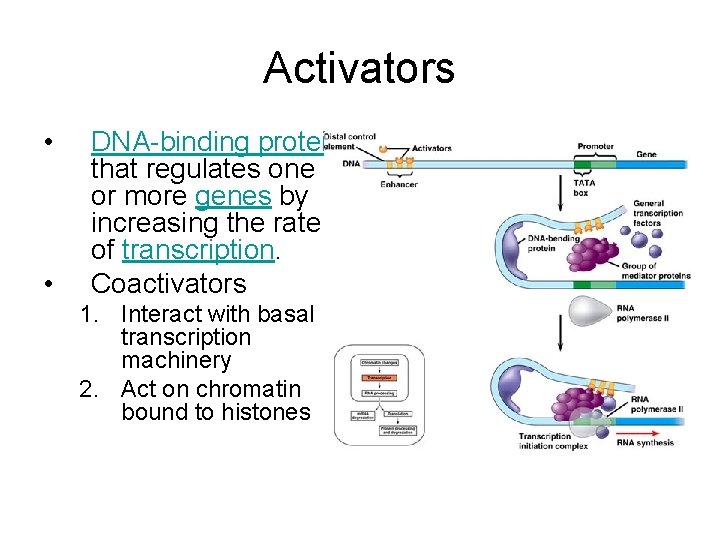
Activators • • DNA-binding protein that regulates one or more genes by increasing the rate of transcription. Coactivators 1. Interact with basal transcription machinery 2. Act on chromatin bound to histones

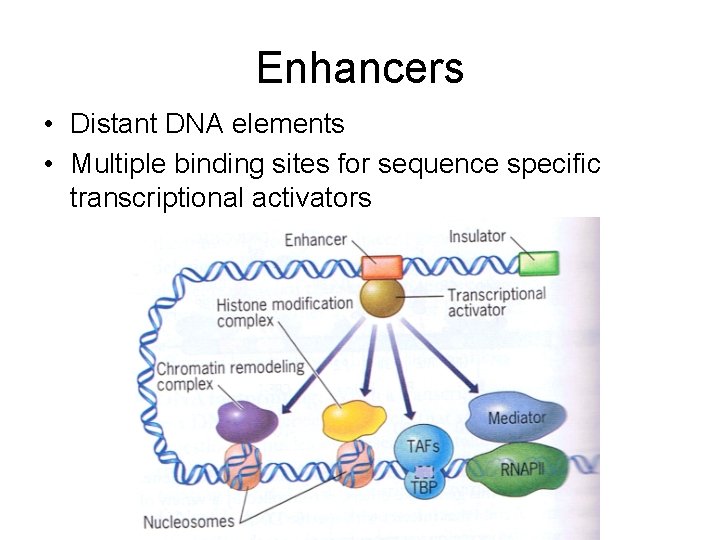
Enhancers • Distant DNA elements • Multiple binding sites for sequence specific transcriptional activators

Translational Control • m. RNA localisation • m. RNA translation • m. RNA stability

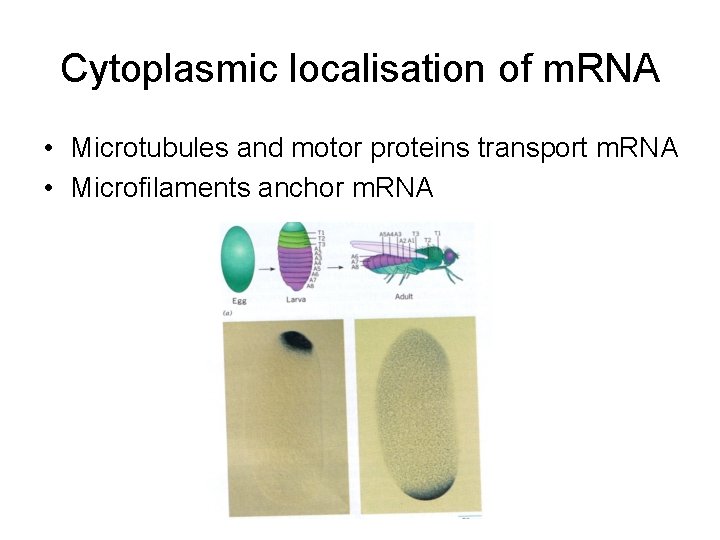
Cytoplasmic localisation of m. RNA • Microtubules and motor proteins transport m. RNA • Microfilaments anchor m. RNA

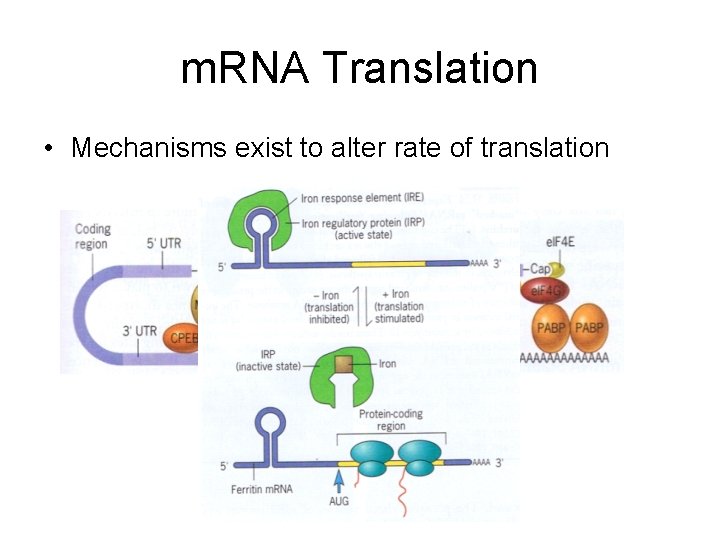
m. RNA Translation • Mechanisms exist to alter rate of translation

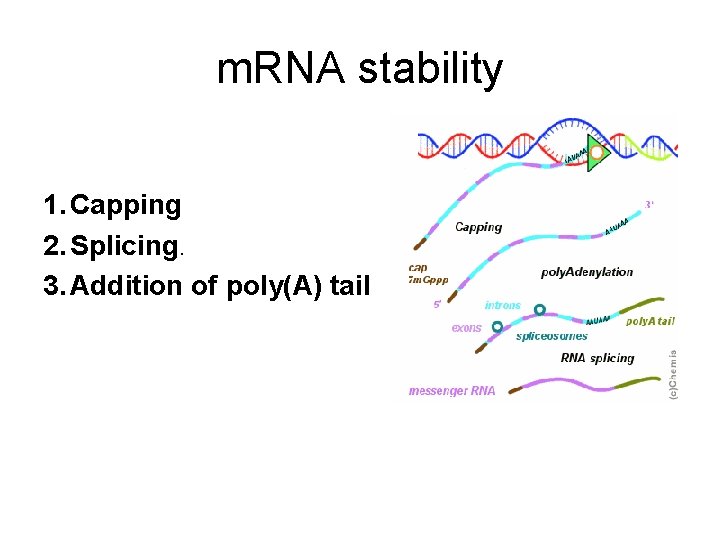
m. RNA stability 1. Capping 2. Splicing. 3. Addition of poly(A) tail




Splicing • removes the introns, noncoding regions that are transcribed into RNA, in order to make the m. RNA able to create proteins. • Cells do this by spliceosome's binding on either side of an intron, looping the intron into a circle and then cleaving it off. • The two ends of the exons are then joined together


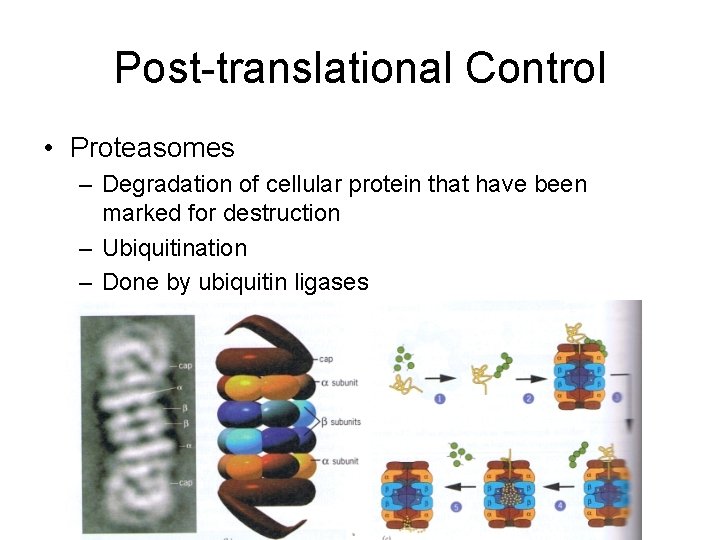
Post-translational Control • Proteasomes – Degradation of cellular protein that have been marked for destruction – Ubiquitination – Done by ubiquitin ligases
 Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Chapter 18
Chapter 18 Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Regulation of gene expression
Regulation of gene expression Regulation of gene expression in bacteria
Regulation of gene expression in bacteria Chapter 17 gene expression from gene to protein
Chapter 17 gene expression from gene to protein Chapter 12 section 4 gene regulation and mutations
Chapter 12 section 4 gene regulation and mutations What is negative control
What is negative control Gene structure prokaryotes vs eukaryotes
Gene structure prokaryotes vs eukaryotes Section 12-1 dna answers
Section 12-1 dna answers Section 12-5 gene regulation
Section 12-5 gene regulation What is gene regulation
What is gene regulation Prokaryotes vs eukaryotes gene regulation
Prokaryotes vs eukaryotes gene regulation Differential gene regulation
Differential gene regulation Gene regulation
Gene regulation Section 4 gene regulation and mutation
Section 4 gene regulation and mutation Gene by gene test results
Gene by gene test results Gene expression omnibus tutorial
Gene expression omnibus tutorial Gene expression
Gene expression What is the first step in gene expression
What is the first step in gene expression Gene expression
Gene expression