PURINE SYNTHESIS Synthesis of PURINE nucleotides AMP GMP




- Slides: 40

PURINE SYNTHESIS

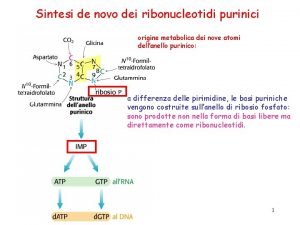
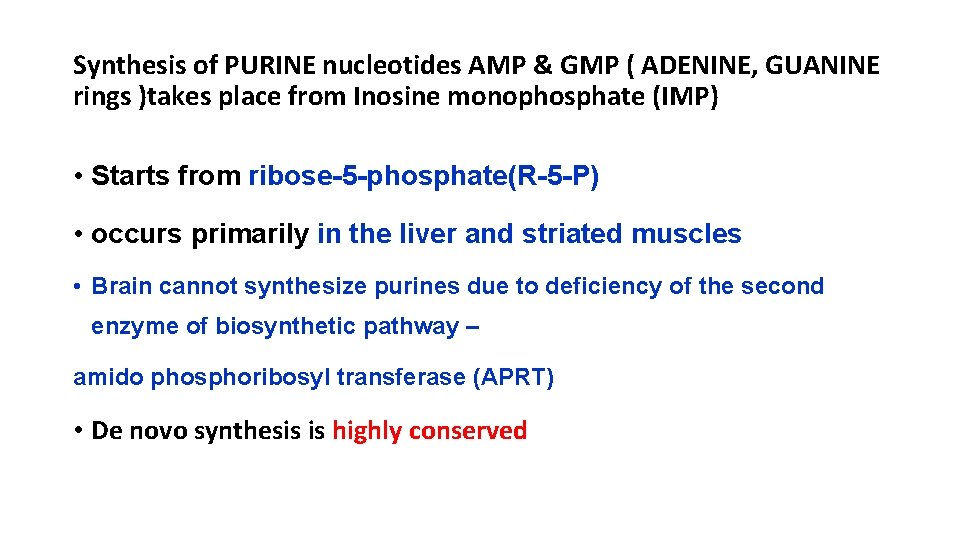
Synthesis of PURINE nucleotides AMP & GMP ( ADENINE, GUANINE rings )takes place from Inosine monophosphate (IMP) • Starts from ribose-5 -phosphate(R-5 -P) • occurs primarily in the liver and striated muscles • Brain cannot synthesize purines due to deficiency of the second enzyme of biosynthetic pathway – amido phosphoribosyl transferase (APRT) • De novo synthesis is highly conserved

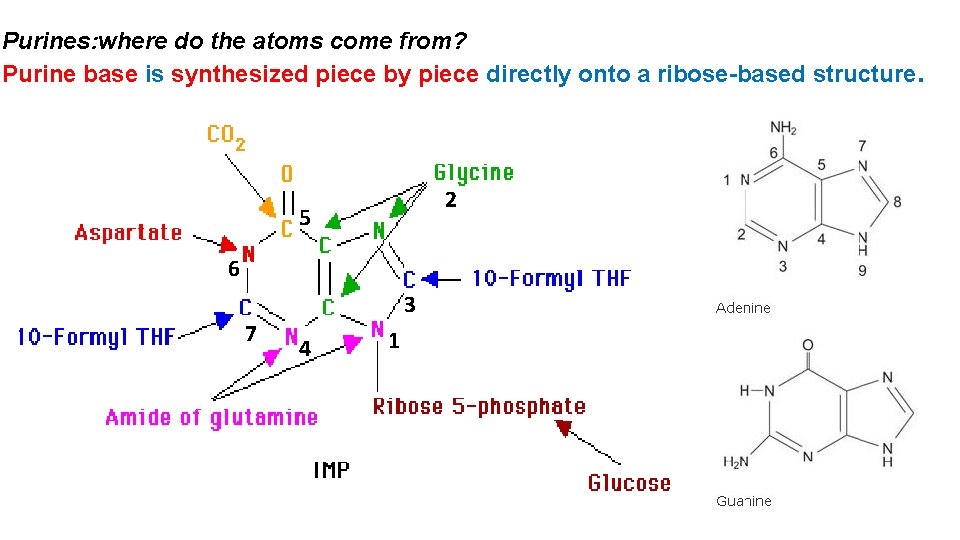
Purines: where do the atoms come from? Purine base is synthesized piece by piece directly onto a ribose-based structure. 2 5 6 7 3 4 1

Purine: De novo synthesis • Site: In cytosol of liver, small intestine and thymus • Characteristics: • Purines are synthesized using 5 -phosphoribose(R-5 -P) as the starting material. • PRPP(5 -phosphoribosyl-1 -pyrophosphate) is active donor of R-5 -P.

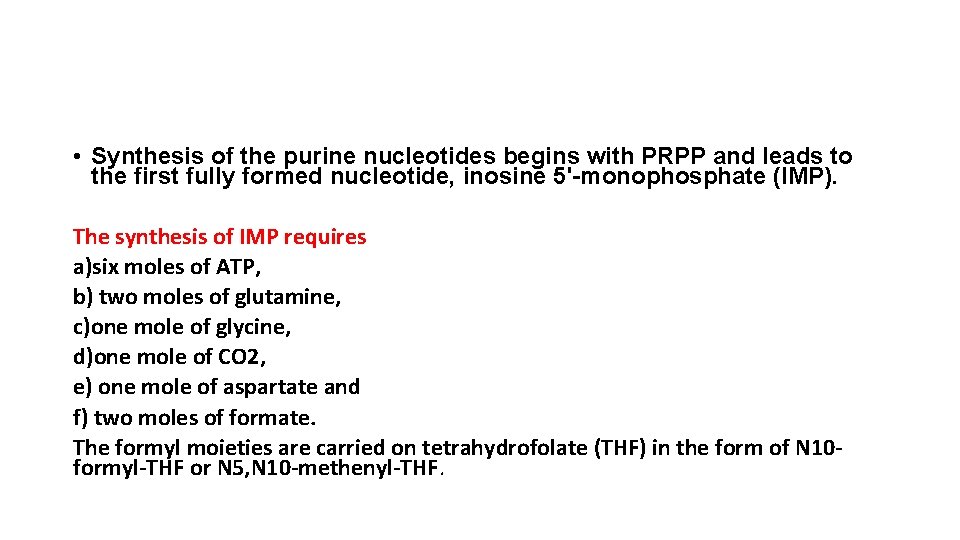
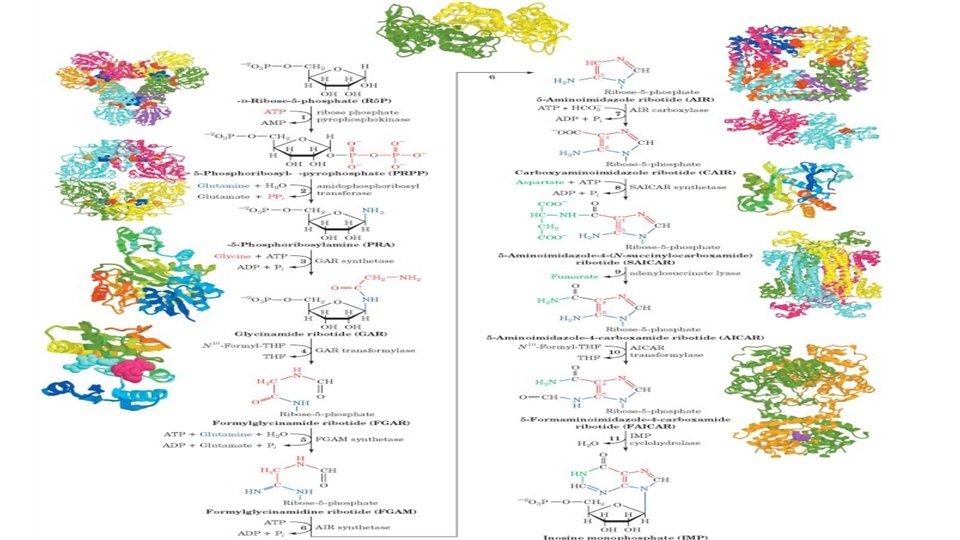
• Synthesis of the purine nucleotides begins with PRPP and leads to the first fully formed nucleotide, inosine 5'-monophosphate (IMP). The synthesis of IMP requires a)six moles of ATP, b) two moles of glutamine, c)one mole of glycine, d)one mole of CO 2, e) one mole of aspartate and f) two moles of formate. The formyl moieties are carried on tetrahydrofolate (THF) in the form of N 10 formyl-THF or N 5, N 10 -methenyl-THF.

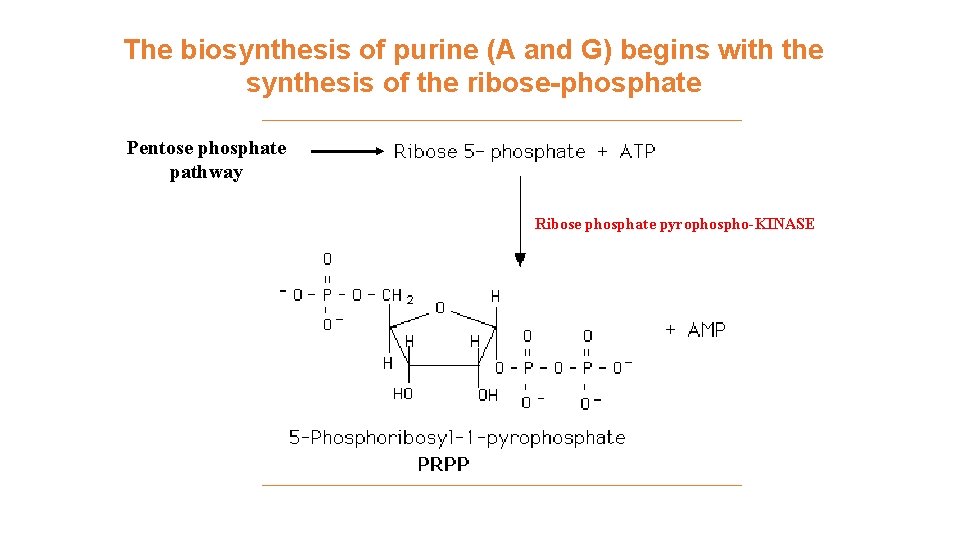
The biosynthesis of purine (A and G) begins with the synthesis of the ribose-phosphate Pentose phosphate pathway Ribose phosphate pyrophospho-KINASE

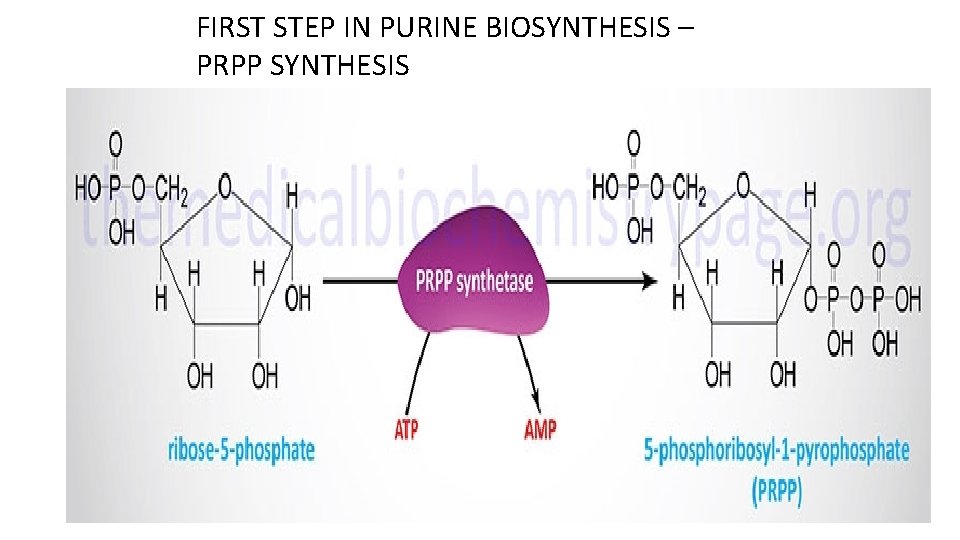
FIRST STEP IN PURINE BIOSYNTHESIS – PRPP SYNTHESIS

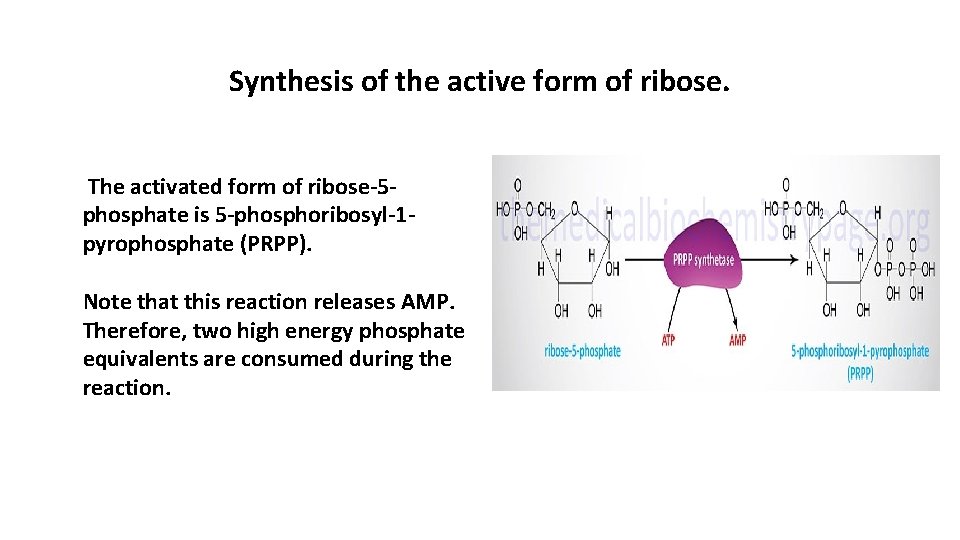
Synthesis of the active form of ribose. The activated form of ribose-5 phosphate is 5 -phosphoribosyl-1 pyrophosphate (PRPP). Note that this reaction releases AMP. Therefore, two high energy phosphate equivalents are consumed during the reaction.

Individual Steps

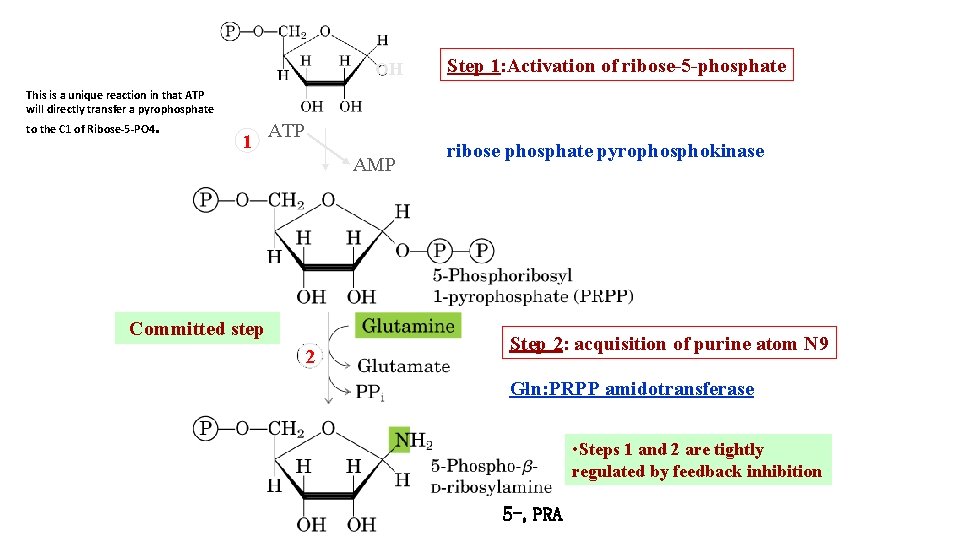
OH Step 1: Activation of ribose-5 -phosphate This is a unique reaction in that ATP will directly transfer a pyrophosphate to the C 1 of Ribose-5 -PO 4. 1 ATP AMP Committed step 2 ribose phosphate pyrophosphokinase Step 2: acquisition of purine atom N 9 Gln: PRPP amidotransferase • Steps 1 and 2 are tightly regulated by feedback inhibition 5 -, PRA

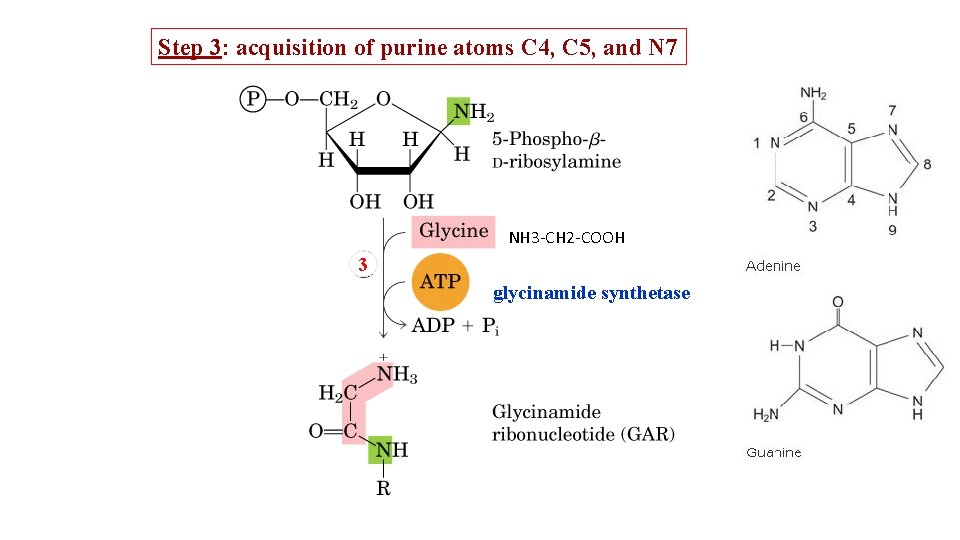
Step 3: acquisition of purine atoms C 4, C 5, and N 7 NH 3 -CH 2 -COOH 3 glycinamide synthetase

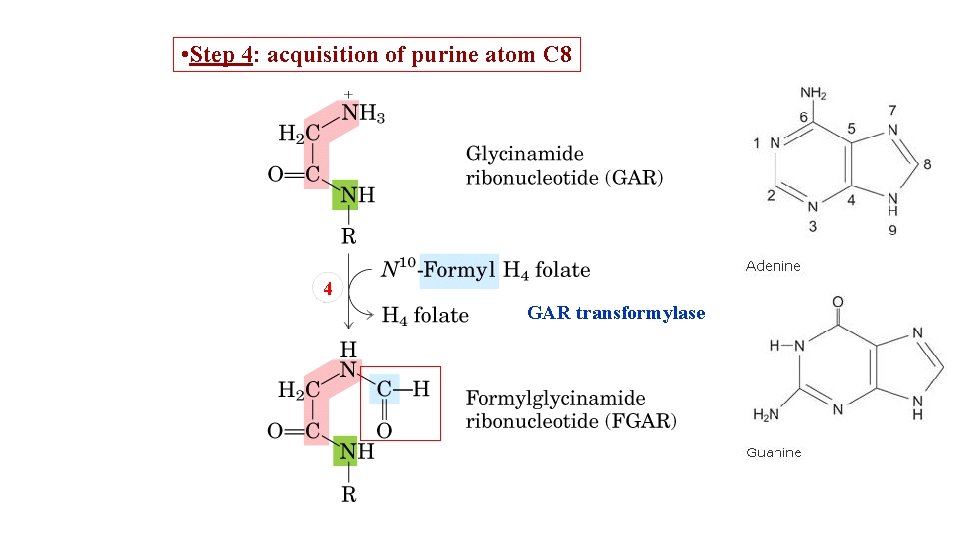
• Step 4: acquisition of purine atom C 8 4 GAR transformylase

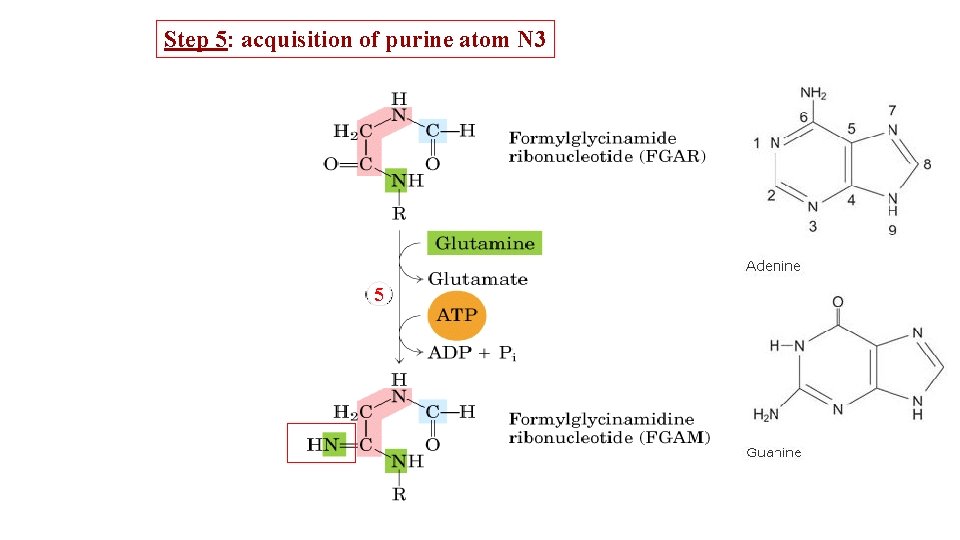
Step 5: acquisition of purine atom N 3 5

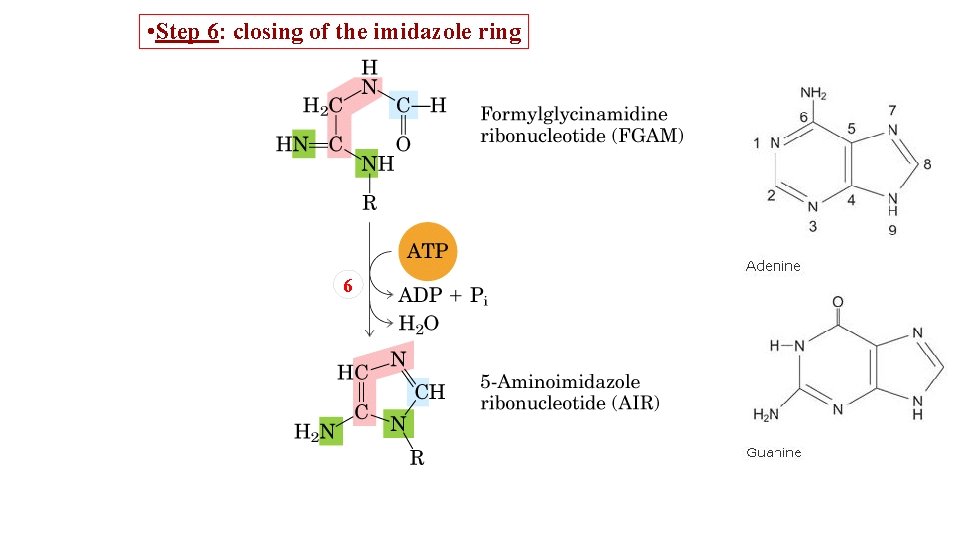
• Step 6: closing of the imidazole ring 6

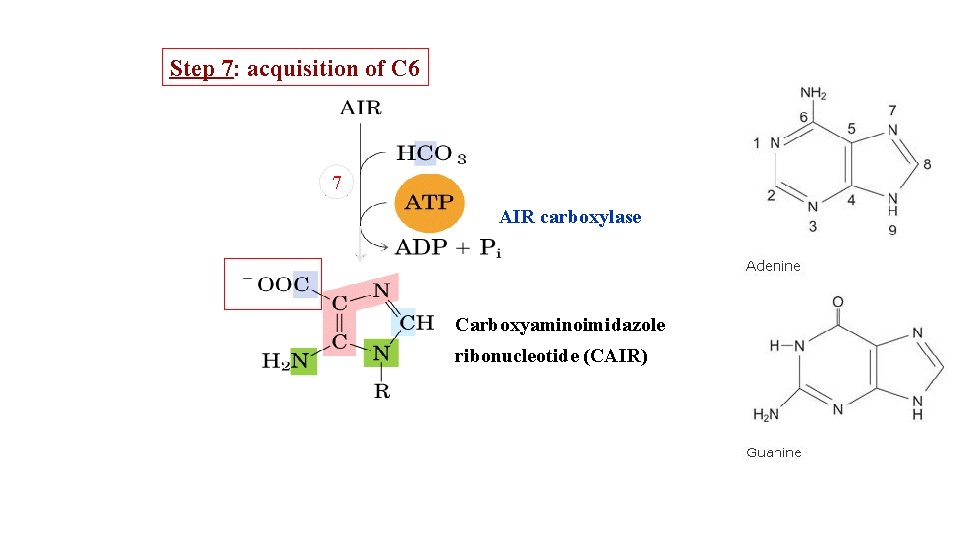
Step 7: acquisition of C 6 7 AIR carboxylase Carboxyaminoimidazole ribonucleotide (CAIR)

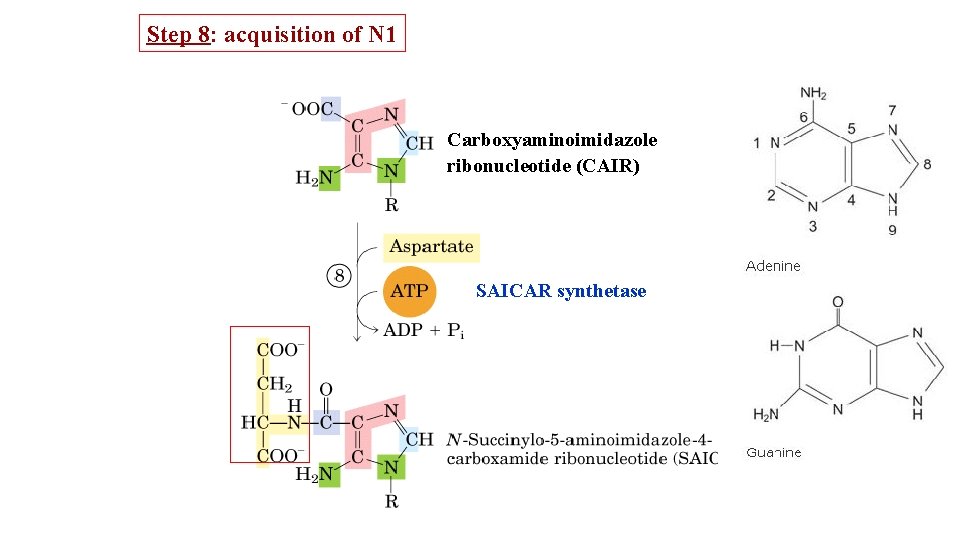
Step 8: acquisition of N 1 Carboxyaminoimidazole ribonucleotide (CAIR) SAICAR synthetase

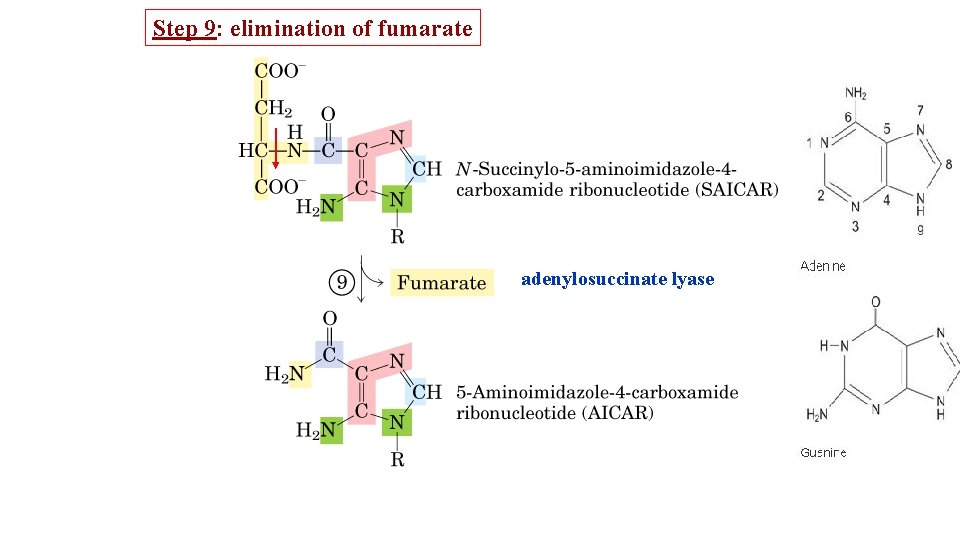
Step 9: elimination of fumarate adenylosuccinate lyase

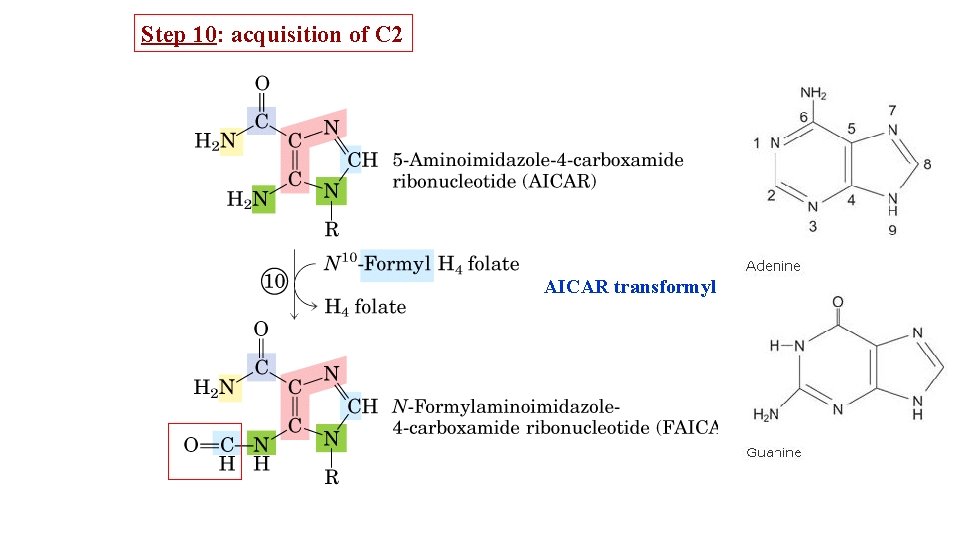
Step 10: acquisition of C 2 AICAR transformylase

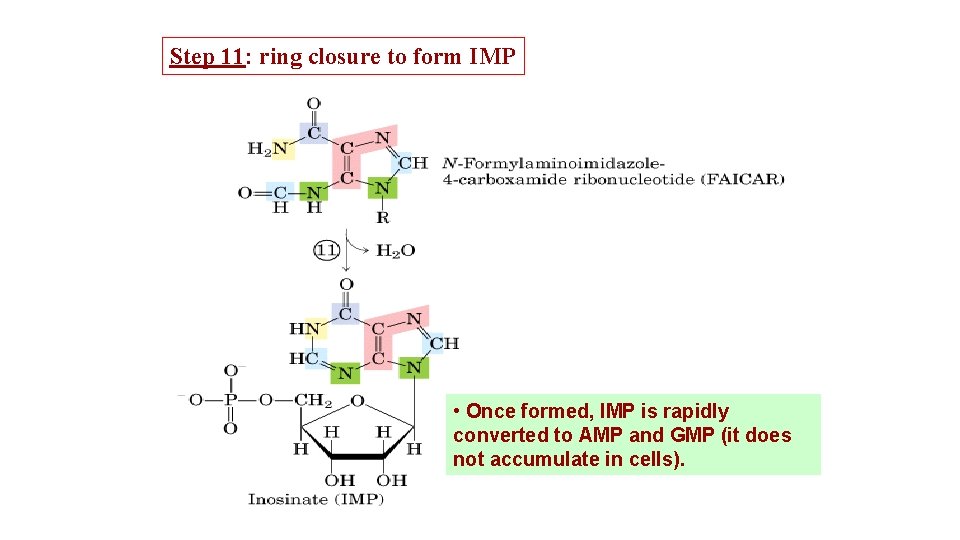
Step 11: ring closure to form IMP • Once formed, IMP is rapidly converted to AMP and GMP (it does not accumulate in cells).

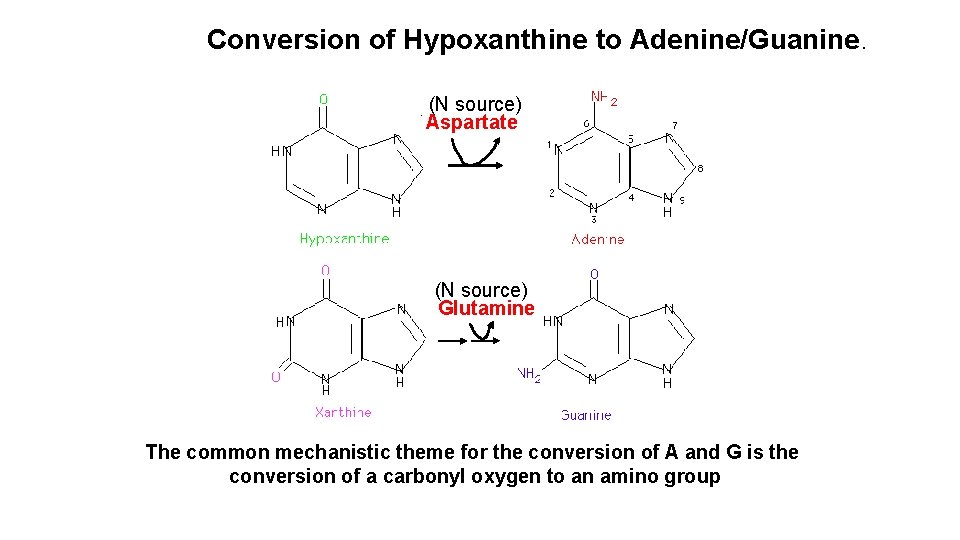
Conversion of Hypoxanthine to Adenine/Guanine. (N source) Aspartate (N source) Glutamine The common mechanistic theme for the conversion of A and G is the conversion of a carbonyl oxygen to an amino group


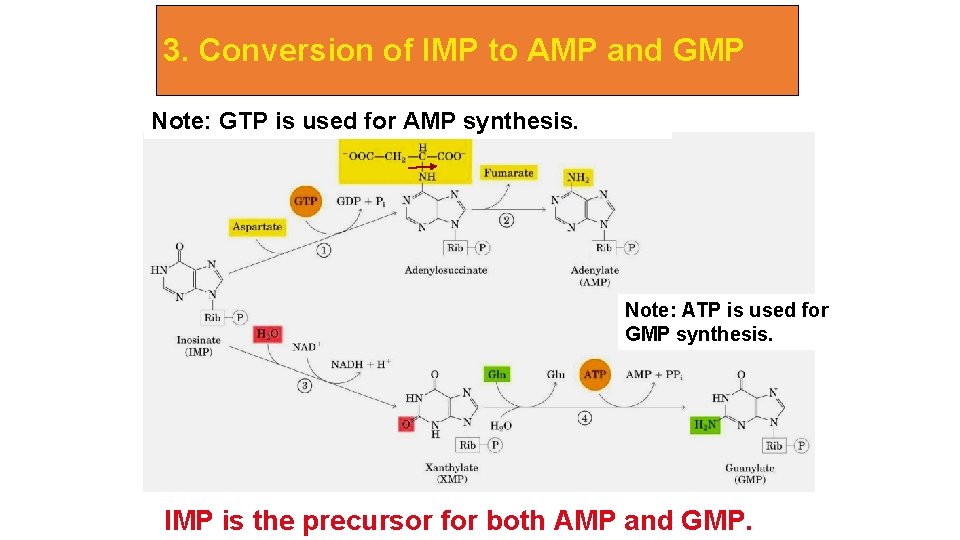
3. Conversion of IMP to AMP and GMP Note: GTP is used for AMP synthesis. Note: ATP is used for GMP synthesis. IMP is the precursor for both AMP and GMP.

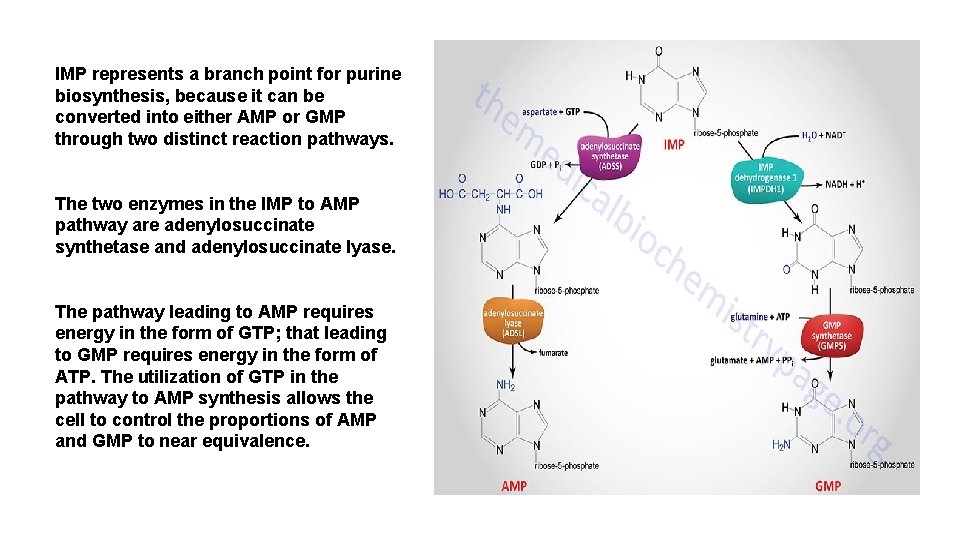
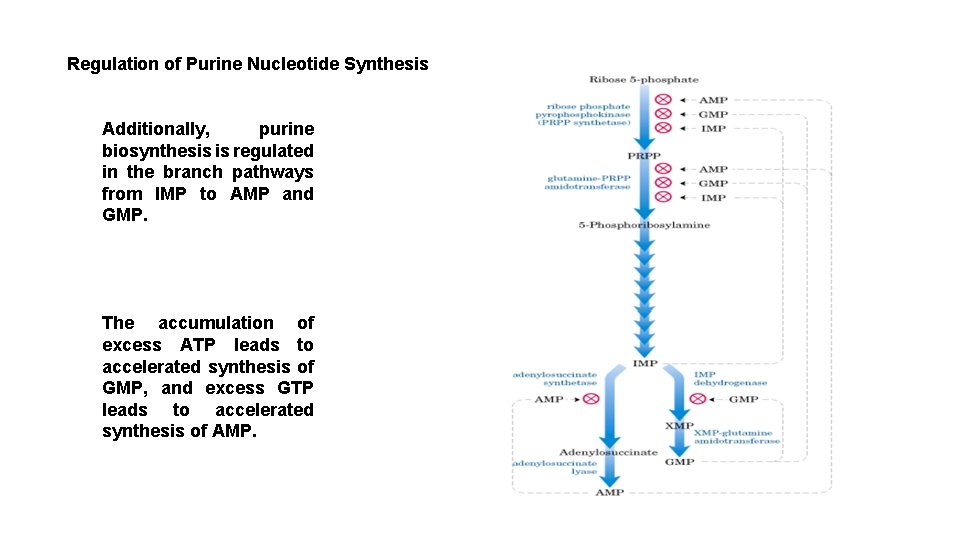
IMP represents a branch point for purine biosynthesis, because it can be converted into either AMP or GMP through two distinct reaction pathways. The two enzymes in the IMP to AMP pathway are adenylosuccinate synthetase and adenylosuccinate lyase. The pathway leading to AMP requires energy in the form of GTP; that leading to GMP requires energy in the form of ATP. The utilization of GTP in the pathway to AMP synthesis allows the cell to control the proportions of AMP and GMP to near equivalence.

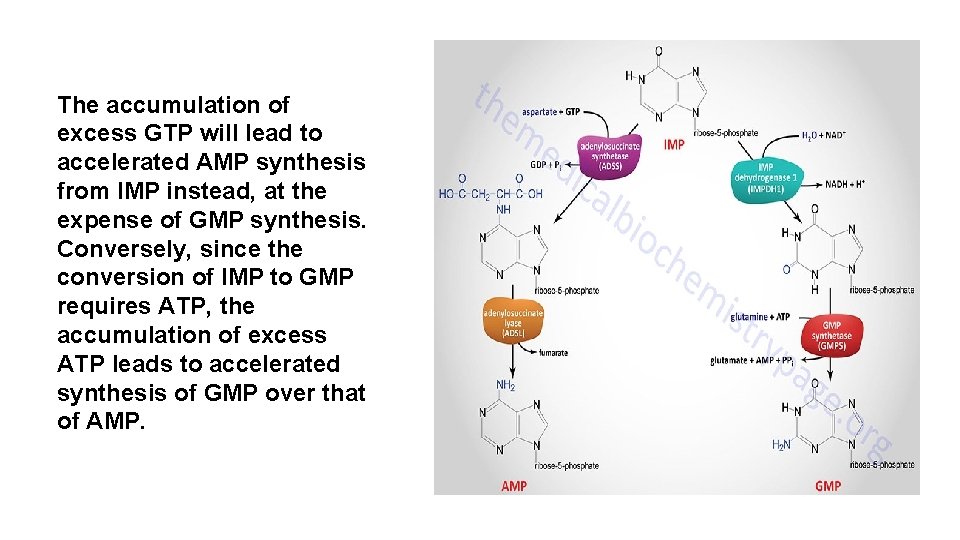
The accumulation of excess GTP will lead to accelerated AMP synthesis from IMP instead, at the expense of GMP synthesis. Conversely, since the conversion of IMP to GMP requires ATP, the accumulation of excess ATP leads to accelerated synthesis of GMP over that of AMP.

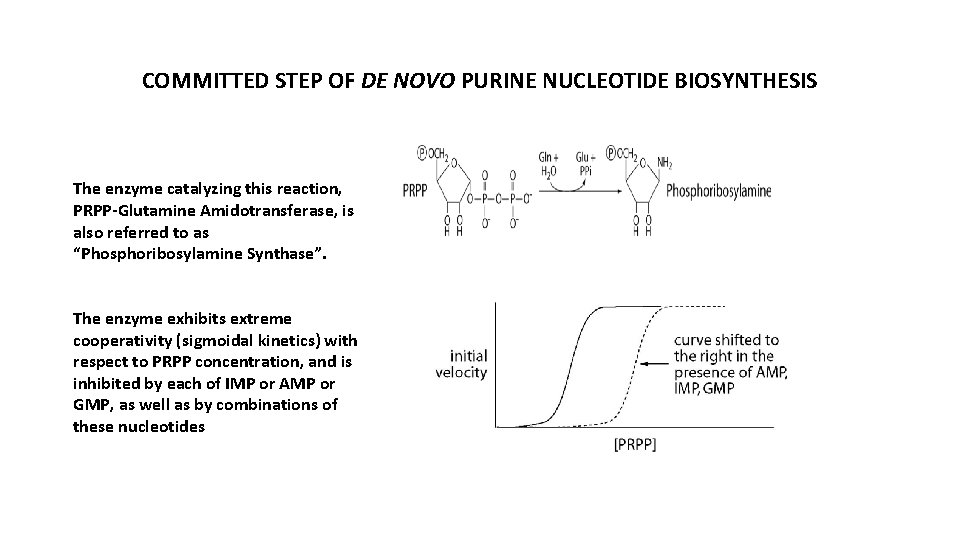
COMMITTED STEP OF DE NOVO PURINE NUCLEOTIDE BIOSYNTHESIS The enzyme catalyzing this reaction, PRPP-Glutamine Amidotransferase, is also referred to as “Phosphoribosylamine Synthase”. The enzyme exhibits extreme cooperativity (sigmoidal kinetics) with respect to PRPP concentration, and is inhibited by each of IMP or AMP or GMP, as well as by combinations of these nucleotides

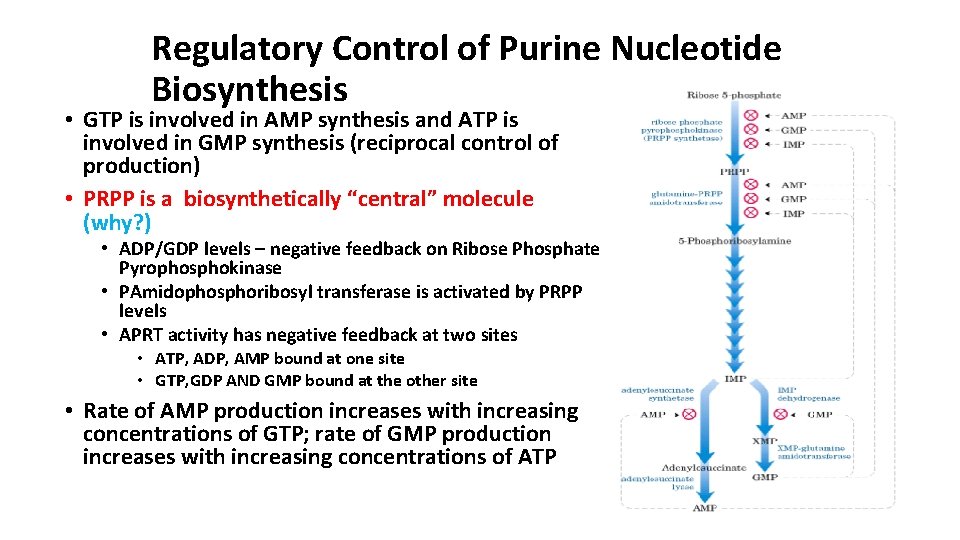
Regulatory Control of Purine Nucleotide Biosynthesis • GTP is involved in AMP synthesis and ATP is involved in GMP synthesis (reciprocal control of production) • PRPP is a biosynthetically “central” molecule (why? ) • ADP/GDP levels – negative feedback on Ribose Phosphate Pyrophosphokinase • PAmidophosphoribosyl transferase is activated by PRPP levels • APRT activity has negative feedback at two sites • ATP, ADP, AMP bound at one site • GTP, GDP AND GMP bound at the other site • Rate of AMP production increases with increasing concentrations of GTP; rate of GMP production increases with increasing concentrations of ATP

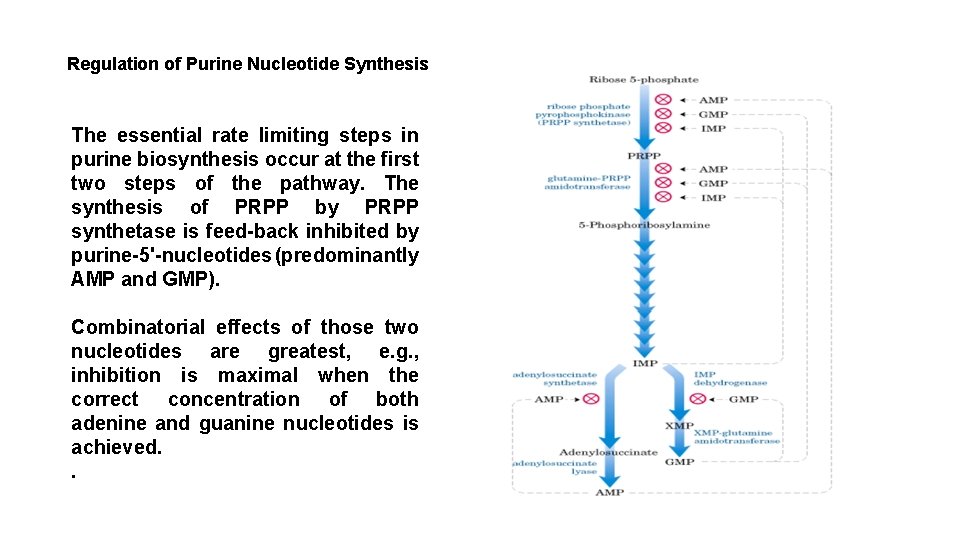
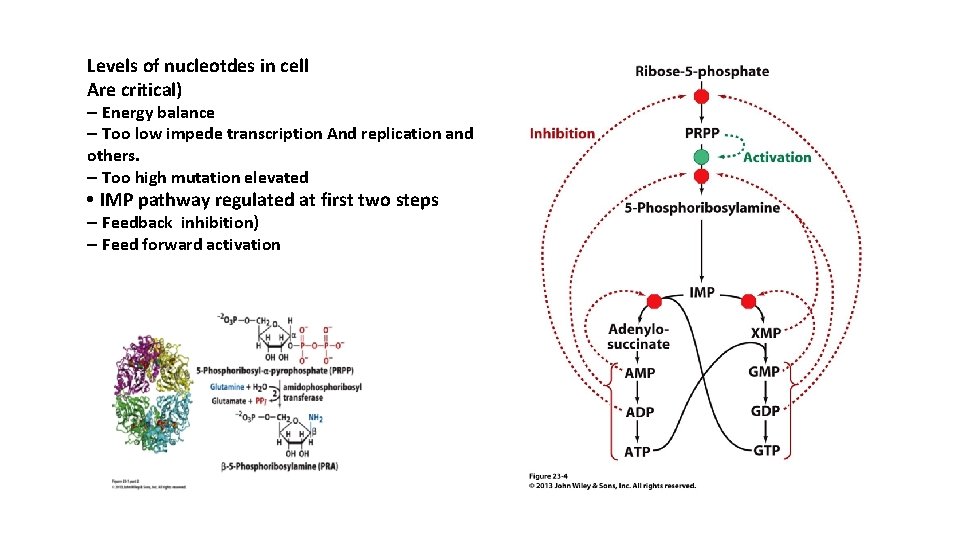
Regulation of Purine Nucleotide Synthesis The essential rate limiting steps in purine biosynthesis occur at the first two steps of the pathway. The synthesis of PRPP by PRPP synthetase is feed-back inhibited by purine-5'-nucleotides (predominantly AMP and GMP). Combinatorial effects of those two nucleotides are greatest, e. g. , inhibition is maximal when the correct concentration of both adenine and guanine nucleotides is achieved. .

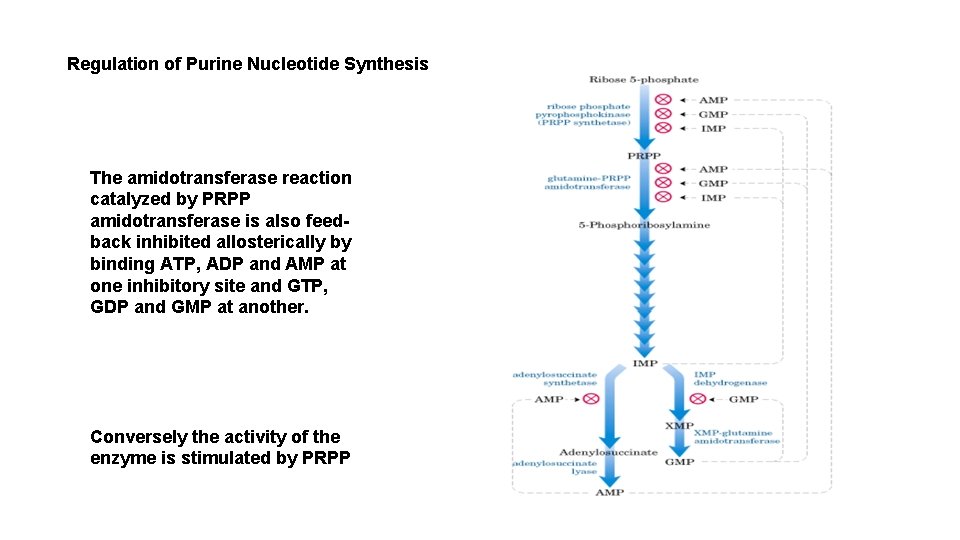
Regulation of Purine Nucleotide Synthesis The amidotransferase reaction catalyzed by PRPP amidotransferase is also feedback inhibited allosterically by binding ATP, ADP and AMP at one inhibitory site and GTP, GDP and GMP at another. Conversely the activity of the enzyme is stimulated by PRPP

Regulation of Purine Nucleotide Synthesis Additionally, purine biosynthesis is regulated in the branch pathways from IMP to AMP and GMP. The accumulation of excess ATP leads to accelerated synthesis of GMP, and excess GTP leads to accelerated synthesis of AMP.

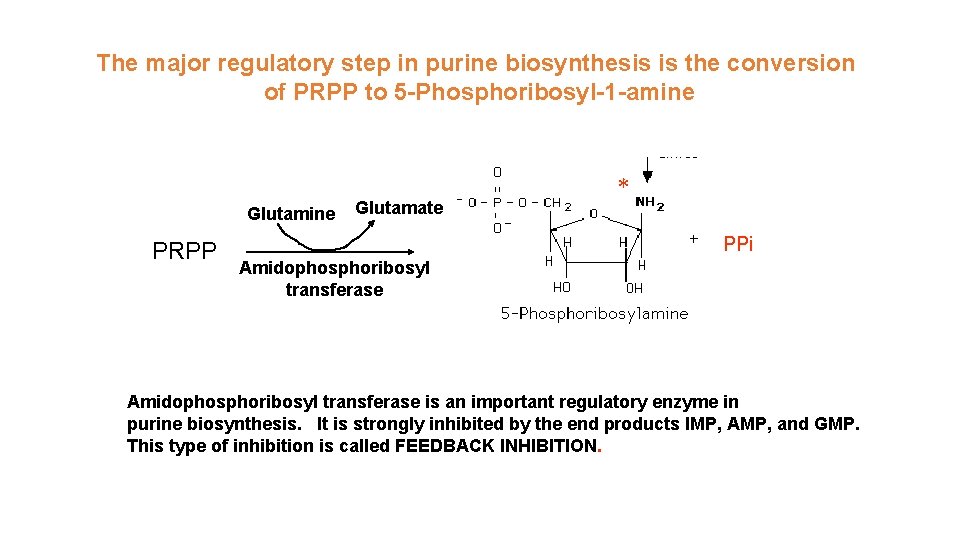
The major regulatory step in purine biosynthesis is the conversion of PRPP to 5 -Phosphoribosyl-1 -amine Glutamine PRPP Glutamate * PPi Amidophosphoribosyl transferase is an important regulatory enzyme in purine biosynthesis. It is strongly inhibited by the end products IMP, AMP, and GMP. This type of inhibition is called FEEDBACK INHIBITION.

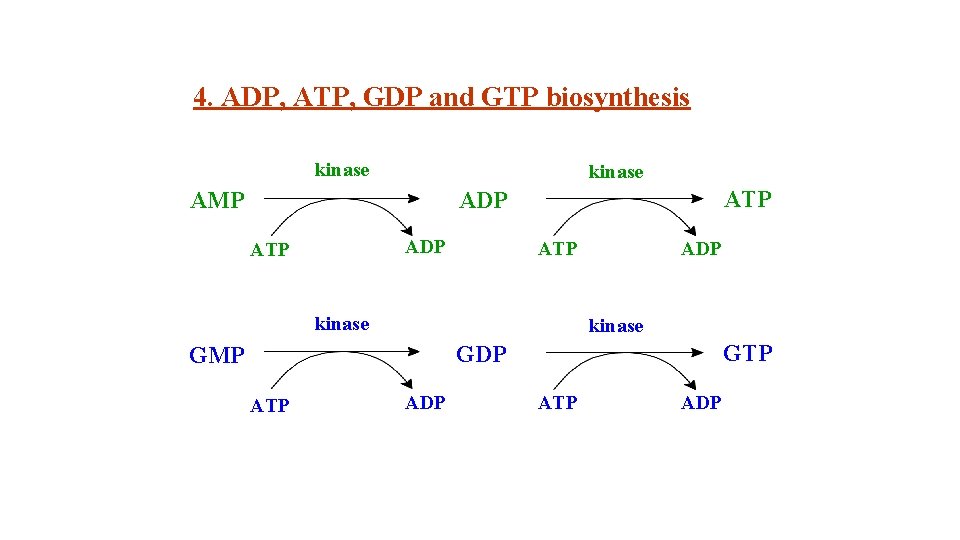
4. ADP, ATP, GDP and GTP biosynthesis kinase AMP ATP ADP ATP kinase ADP kinase GTP GDP GMP ATP ADP

Levels of nucleotdes in cell Are critical) – Energy balance – Too low impede transcription And replication and others. – Too high mutation elevated • IMP pathway regulated at first two steps – Feedback inhibition) – Feed forward activation

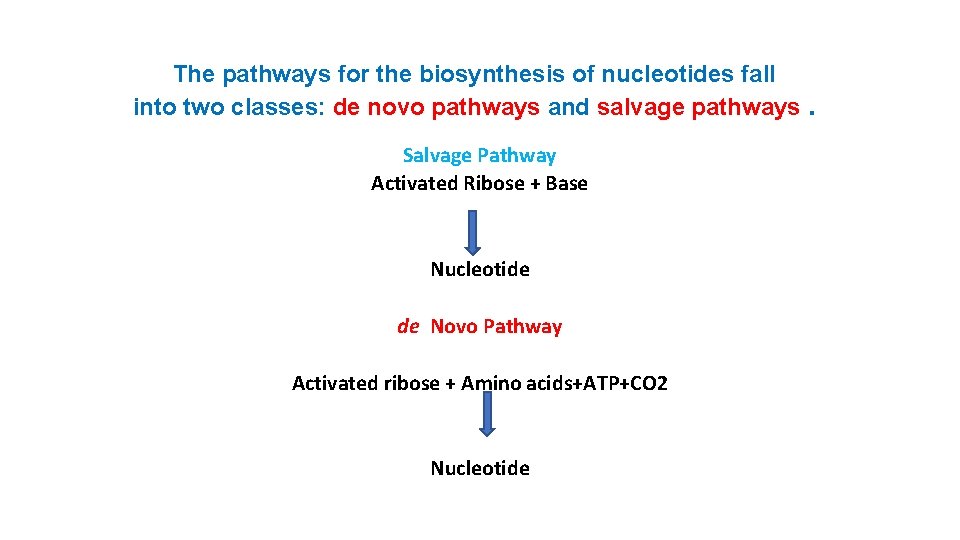
The pathways for the biosynthesis of nucleotides fall into two classes: de novo pathways and salvage pathways. Salvage Pathway Activated Ribose + Base Nucleotide de Novo Pathway Activated ribose + Amino acids+ATP+CO 2 Nucleotide

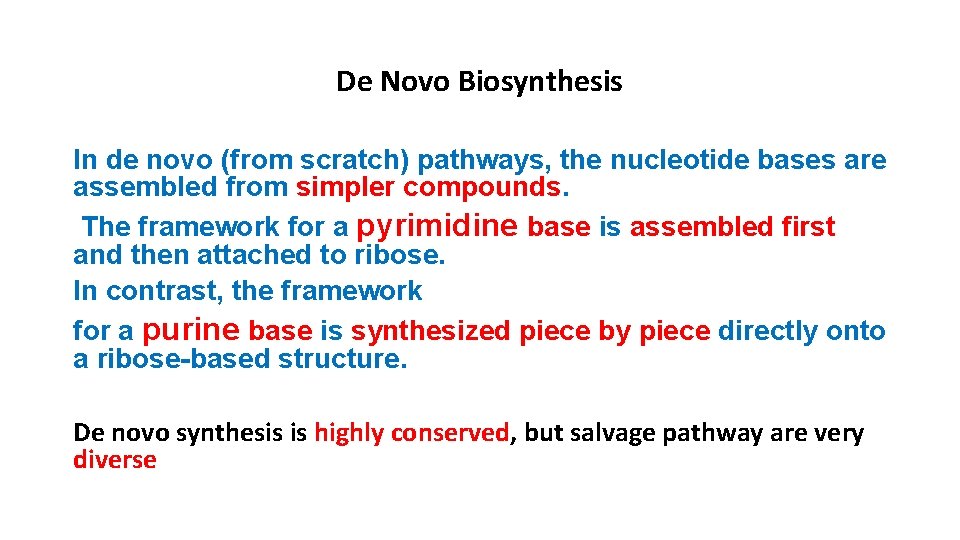
De Novo Biosynthesis In de novo (from scratch) pathways, the nucleotide bases are assembled from simpler compounds. The framework for a pyrimidine base is assembled first and then attached to ribose. In contrast, the framework for a purine base is synthesized piece by piece directly onto a ribose-based structure. De novo synthesis is highly conserved, but salvage pathway are very diverse

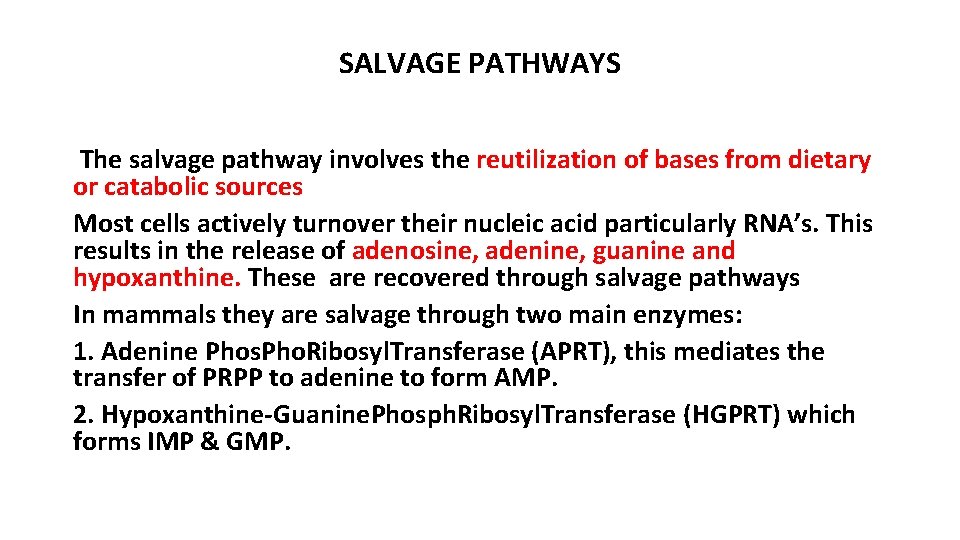
SALVAGE PATHWAYS The salvage pathway involves the reutilization of bases from dietary or catabolic sources Most cells actively turnover their nucleic acid particularly RNA’s. This results in the release of adenosine, adenine, guanine and hypoxanthine. These are recovered through salvage pathways In mammals they are salvage through two main enzymes: 1. Adenine Phos. Pho. Ribosyl. Transferase (APRT), this mediates the transfer of PRPP to adenine to form AMP. 2. Hypoxanthine-Guanine. Phosph. Ribosyl. Transferase (HGPRT) which forms IMP & GMP.

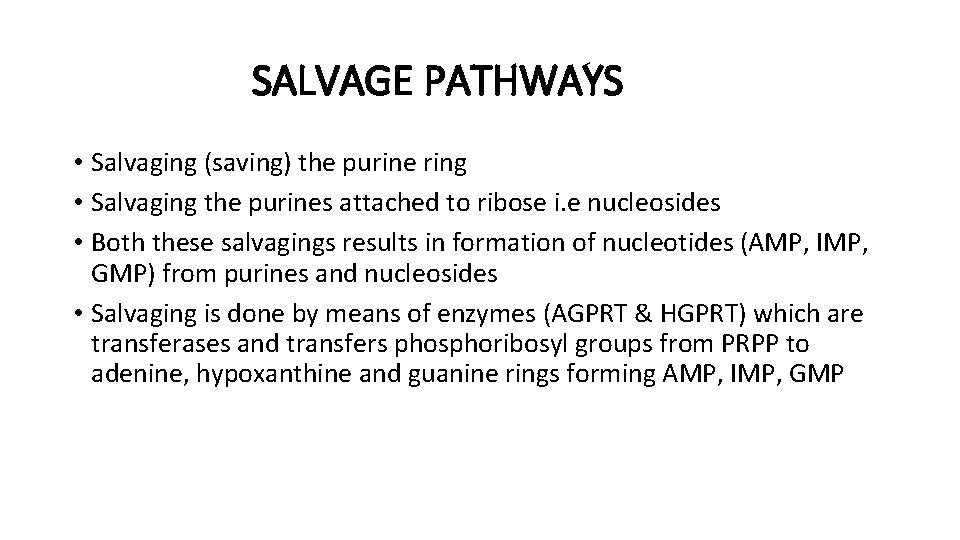
SALVAGE PATHWAYS • Salvaging (saving) the purine ring • Salvaging the purines attached to ribose i. e nucleosides • Both these salvagings results in formation of nucleotides (AMP, IMP, GMP) from purines and nucleosides • Salvaging is done by means of enzymes (AGPRT & HGPRT) which are transferases and transfers phosphoribosyl groups from PRPP to adenine, hypoxanthine and guanine rings forming AMP, IMP, GMP

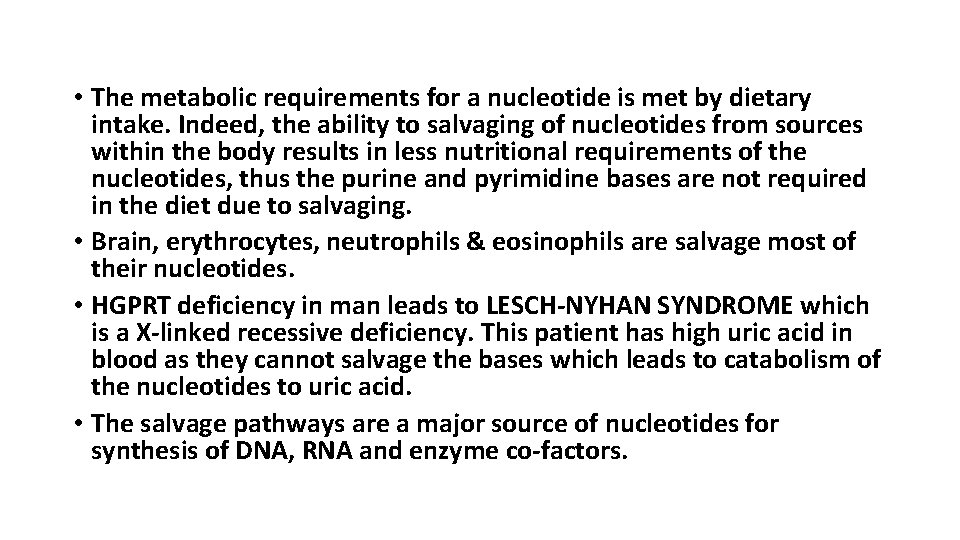
• The metabolic requirements for a nucleotide is met by dietary intake. Indeed, the ability to salvaging of nucleotides from sources within the body results in less nutritional requirements of the nucleotides, thus the purine and pyrimidine bases are not required in the diet due to salvaging. • Brain, erythrocytes, neutrophils & eosinophils are salvage most of their nucleotides. • HGPRT deficiency in man leads to LESCH-NYHAN SYNDROME which is a X-linked recessive deficiency. This patient has high uric acid in blood as they cannot salvage the bases which leads to catabolism of the nucleotides to uric acid. • The salvage pathways are a major source of nucleotides for synthesis of DNA, RNA and enzyme co-factors.

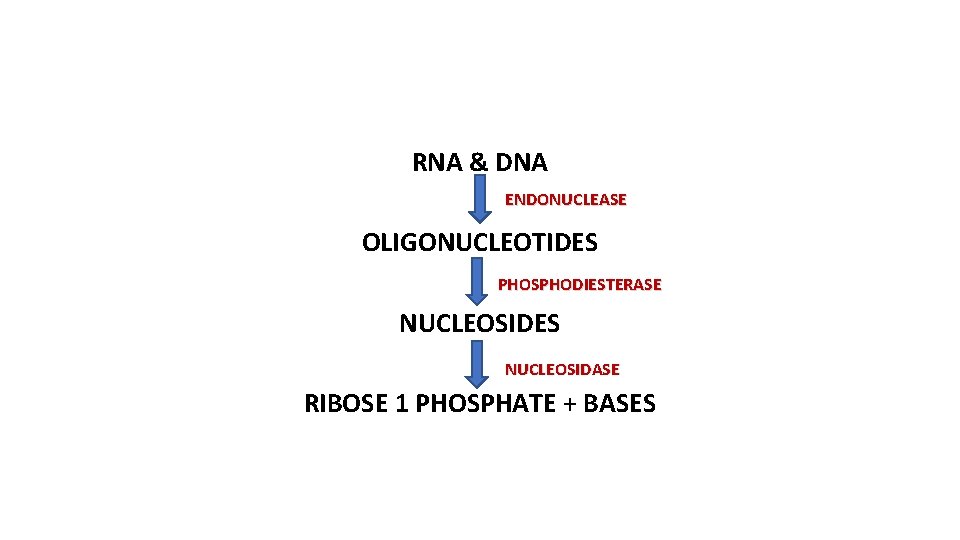
RNA & DNA ENDONUCLEASE OLIGONUCLEOTIDES PHOSPHODIESTERASE NUCLEOSIDES NUCLEOSIDASE RIBOSE 1 PHOSPHATE + BASES

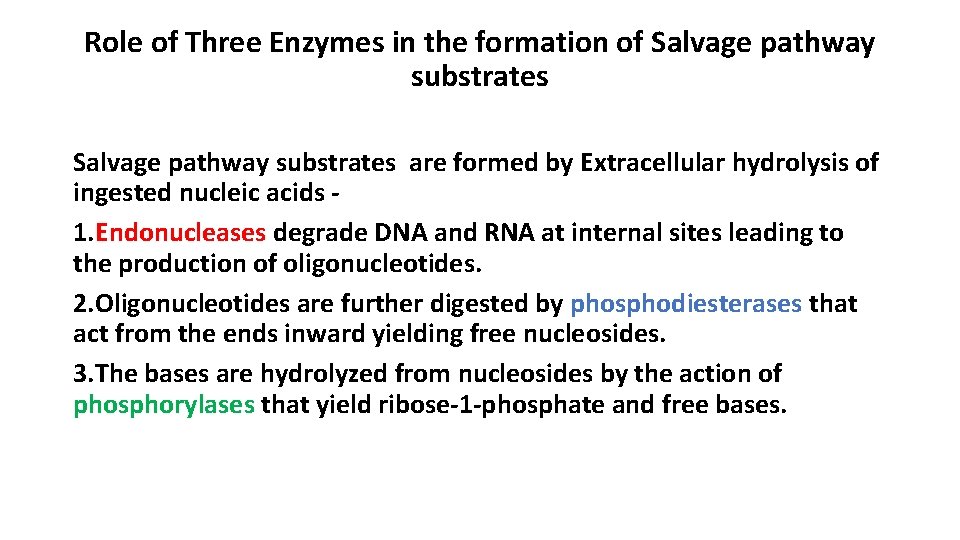
Role of Three Enzymes in the formation of Salvage pathway substrates are formed by Extracellular hydrolysis of ingested nucleic acids 1. Endonucleases degrade DNA and RNA at internal sites leading to the production of oligonucleotides. 2. Oligonucleotides are further digested by phosphodiesterases that act from the ends inward yielding free nucleosides. 3. The bases are hydrolyzed from nucleosides by the action of phosphorylases that yield ribose-1 -phosphate and free bases.

Excess nucleotides If the nucleosides and/or bases are not re-utilized the purine bases are further degraded to uric acid and the pyrimidines to βaminoiosobutyrate, NH 3 and CO 2
 De novo nucleotide synthesis
De novo nucleotide synthesis Amp synthesis
Amp synthesis Difference between ideal op amp and practical op amp
Difference between ideal op amp and practical op amp Order of nucleotides in dna
Order of nucleotides in dna Dna negative charge
Dna negative charge Biomedical importance of nucleotides
Biomedical importance of nucleotides Nucleotides wiki
Nucleotides wiki Nucleotides wiki
Nucleotides wiki Nucleotide to amino acid
Nucleotide to amino acid Pentose sugar in rna
Pentose sugar in rna Dna secret code
Dna secret code Four nucleotides of dna
Four nucleotides of dna When dna untwists, unzips, and nucleotides fill in.
When dna untwists, unzips, and nucleotides fill in. Importance of nucleic acid
Importance of nucleic acid Pyrimidine
Pyrimidine Sintesi de novo purine
Sintesi de novo purine Purine antagonist
Purine antagonist Protoalkaloids
Protoalkaloids Hmp shunt pathway
Hmp shunt pathway Purine bommen
Purine bommen Purine antagonist
Purine antagonist Dna structure labeled hydrogen bonds
Dna structure labeled hydrogen bonds Purine disorders
Purine disorders Formica elica
Formica elica Purine examples
Purine examples Purine alkaloids
Purine alkaloids Purine nucleotide cycle
Purine nucleotide cycle Purine
Purine Prdp biochemistry
Prdp biochemistry Hgprt deficiency
Hgprt deficiency Gmp class d
Gmp class d Components of gmp
Components of gmp Gmp sterile pharmaceutical products
Gmp sterile pharmaceutical products Gmp sterile pharmaceutical products
Gmp sterile pharmaceutical products Objectives of gmp
Objectives of gmp Nafdac gmp guidelines
Nafdac gmp guidelines Principles of gmp
Principles of gmp Target operating model framework
Target operating model framework Chamilo gmp
Chamilo gmp Gmp meaning
Gmp meaning Prerequisite program template
Prerequisite program template