Power in QTL linkage analysis Shaun Purcell Pak
































- Slides: 71

Power in QTL linkage analysis Shaun Purcell & Pak Sham SGDP, Io. P, London, UK F: pshaunpower. ppt

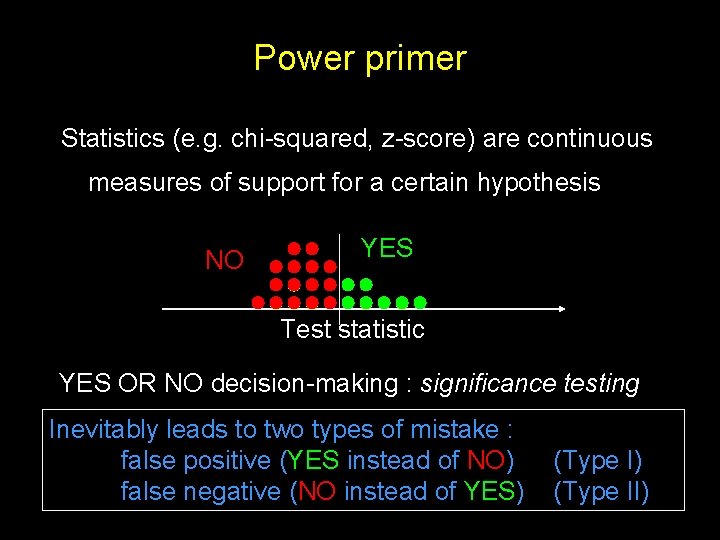
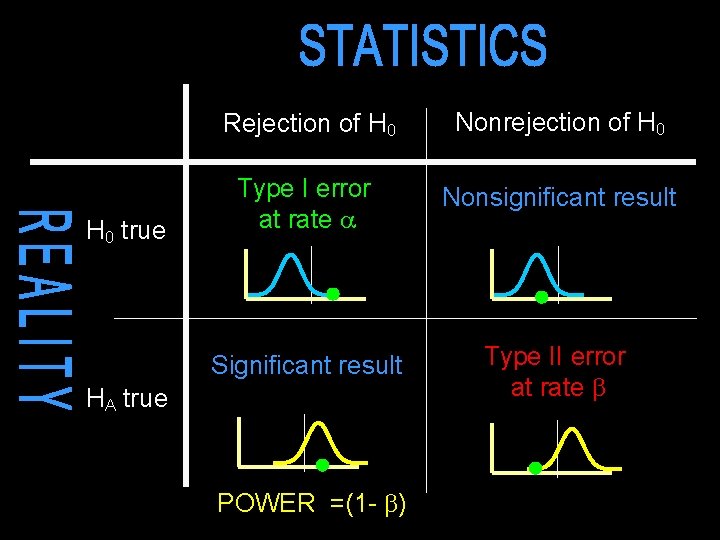
Power primer Statistics (e. g. chi-squared, z-score) are continuous measures of support for a certain hypothesis NO YES Test statistic YES OR NO decision-making : significance testing Inevitably leads to two types of mistake : false positive (YES instead of NO) false negative (NO instead of YES) (Type II)

Hypothesis testing Null hypothesis : no effect A ‘significant’ result means that we can reject the null hypothesis A ‘nonsignificant’ result means that we cannot reject the null hypothesis

Statistical significance The ‘p-value’ The probability of a false positive error if the null were in fact true Typically, we are willing to incorrectly reject the null 5% or 1% of the time (Type I error)

Misunderstandings p - VALUES that the p value is the probability of the null hypothesis being true that high p values mean large and important effects NULL HYPOTHESIS that nonrejection of the null implies its truth

Limitations IF A RESULT IS SIGNIFICANT leads to the conclusion that the null is false BUT, this may be trivial IF A RESULT IS NONSIGNIFICANT leads only to the conclusion that it cannot be concluded that the null is false

Alternate hypothesis Neyman & Pearson (1928) ALTERNATE HYPOTHESIS specifies a precise, non-null state of affairs with associated risk of error

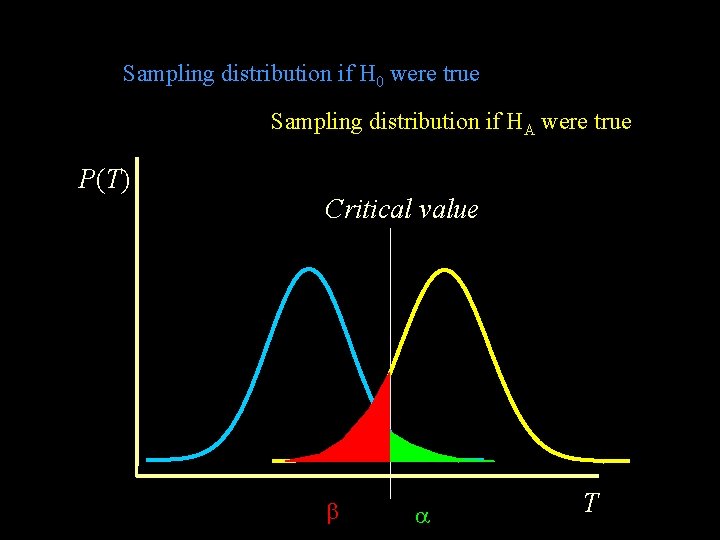
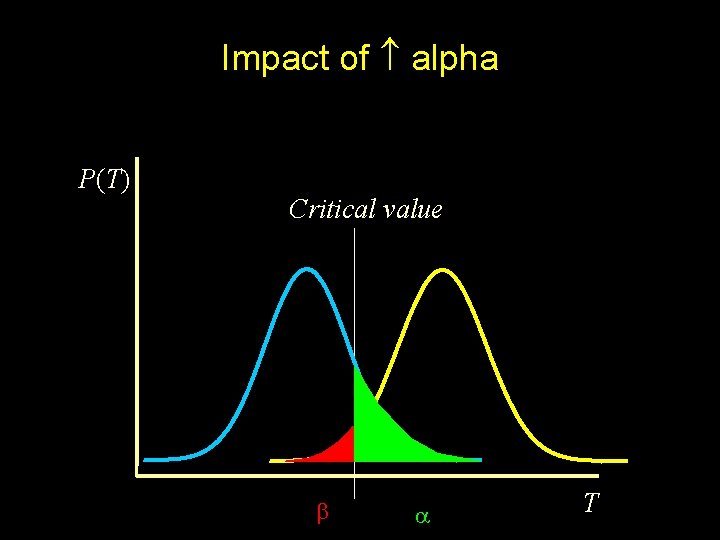
Sampling distribution if H 0 were true Sampling distribution if HA were true P(T) Critical value T

H 0 true Rejection of H 0 Nonrejection of H 0 Type I error at rate Nonsignificant result Significant result Type II error at rate HA true POWER =(1 - )

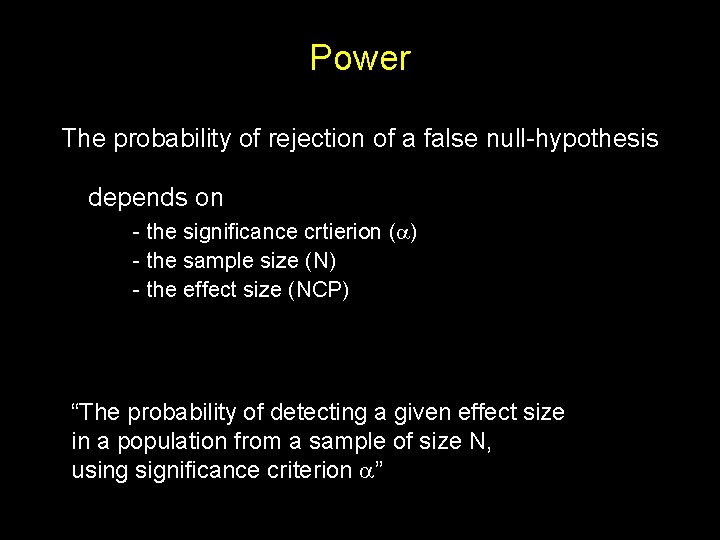
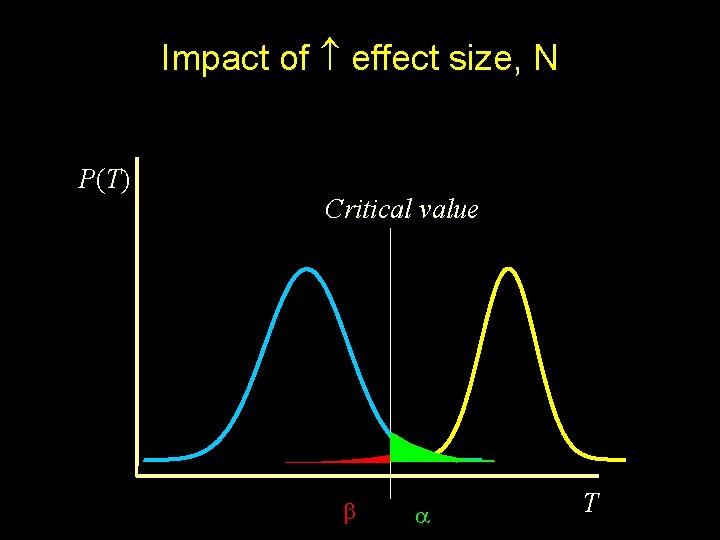
Power The probability of rejection of a false null-hypothesis depends on - the significance crtierion ( ) - the sample size (N) - the effect size (NCP) “The probability of detecting a given effect size in a population from a sample of size N, using significance criterion ”

Impact of alpha P(T) Critical value T

Impact of effect size, N P(T) Critical value T

Applications EXPERIMENTAL DESIGN - avoiding false positives vs. dealing with false negatives MAGNITUDE VS. SIGNIFICANCE - highly significant very important INTERPRETING NONSIGIFICANT RESULTS - nonsignficant results only meaningful if power is high POWER SURVEYS / META-ANALYSES - low power undermines the confidence that can be placed in statistically significant results

Practical Exercise 1 Calculation of power for simple case-control association study. DATA : allele frequency of “A” allele for cases and controls TEST : 2 -by-2 contingency table : chi-squared (1 degree of freedom)

Step 1 : determine expected chi-squared Hypothetical allele frequencies Cases P(A) = 0. 68 Controls P(A) = 0. 54 Sample 150 cases, 150 controls Excel spreadsheet : faculty drive: pshaunchisq. xls Chi-squared statistic = 12. 36

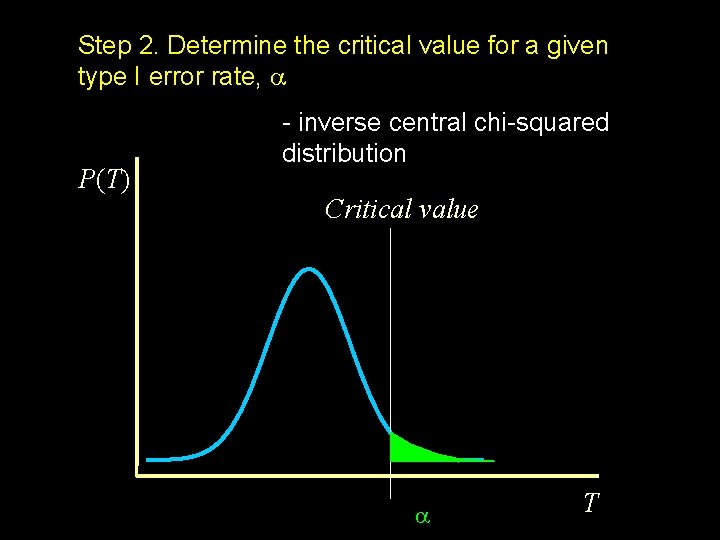
Step 2. Determine the critical value for a given type I error rate, P(T) - inverse central chi-squared distribution Critical value T

http: //workshop. colorado. edu/~pshaun/gpc/pdf. html df = 1 , NCP = 0 X 0. 05 3. 84146 0. 01 6. 63489 0. 001 10. 82754

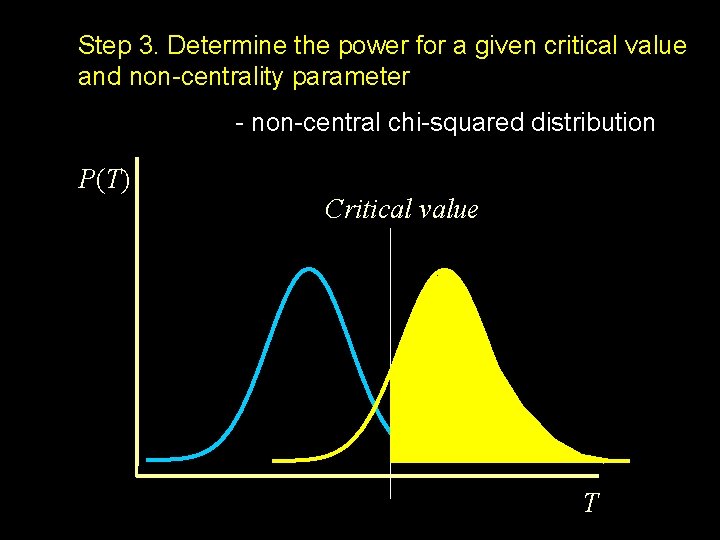
Step 3. Determine the power for a given critical value and non-centrality parameter - non-central chi-squared distribution P(T) Critical value T

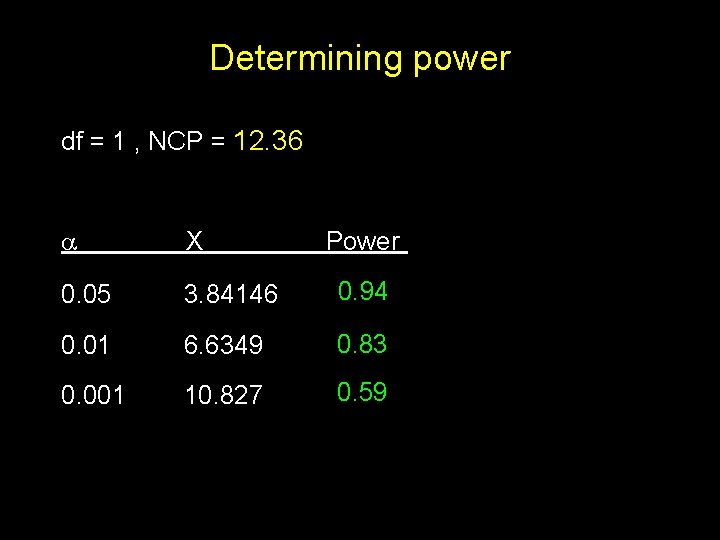
Determining power df = 1 , NCP = 12. 36 X 0. 05 3. 84146 0. 94 0. 01 6. 6349 0. 83 0. 001 10. 827 0. 59 Power

Exercises Using the spreadsheet and the chi-squared calculator, what is power (for the 3 levels of alpha) 1. … if the sample size were 300 for each group? 2. … if allele frequencies were 0. 24 and 0. 18 for 750 cases and 750 controls?

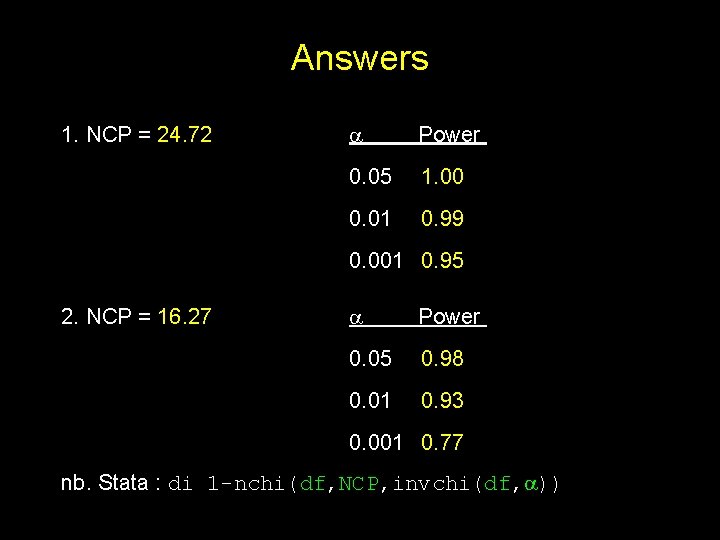
Answers 1. NCP = 24. 72 Power 0. 05 1. 00 0. 01 0. 99 0. 001 0. 95 2. NCP = 16. 27 Power 0. 05 0. 98 0. 01 0. 93 0. 001 0. 77 nb. Stata : di 1 -nchi(df, NCP, invchi(df, ))

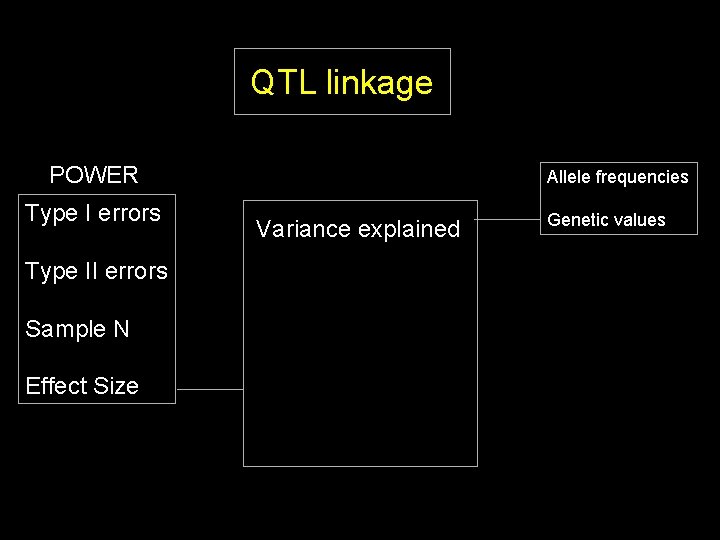
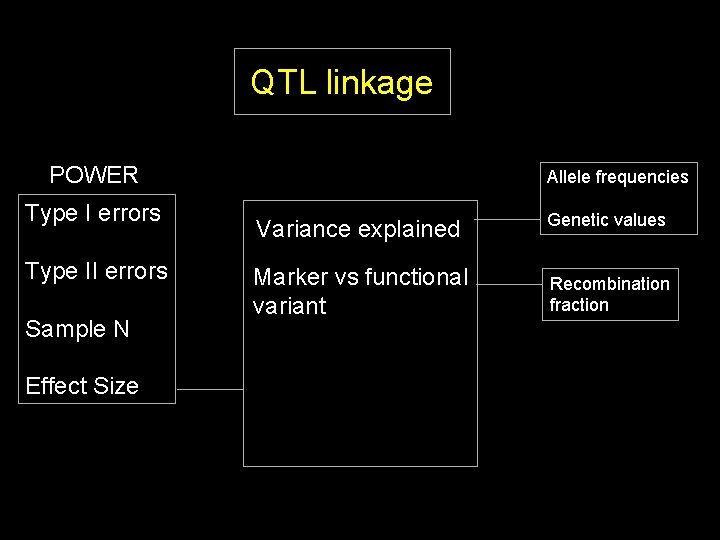
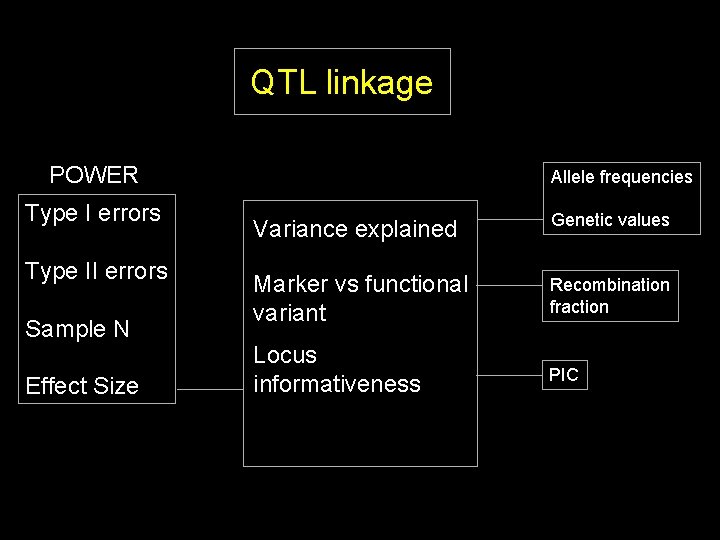
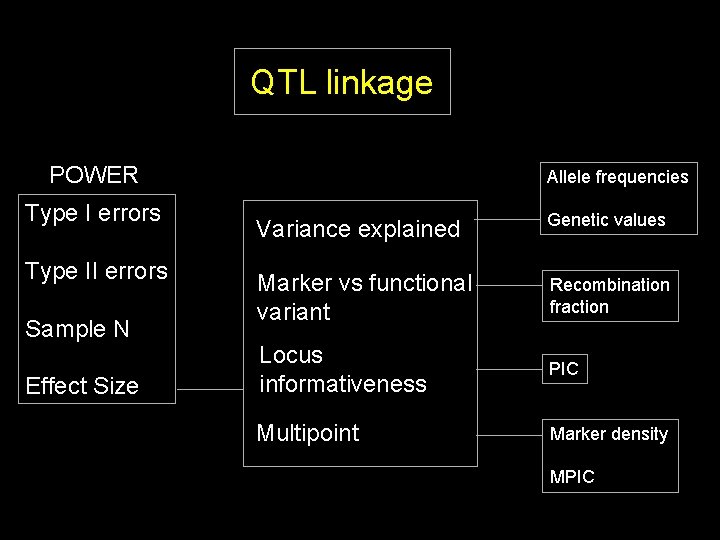
QTL linkage POWER Type I errors Type II errors Sample N Effect Size Allele frequencies Variance explained Genetic values

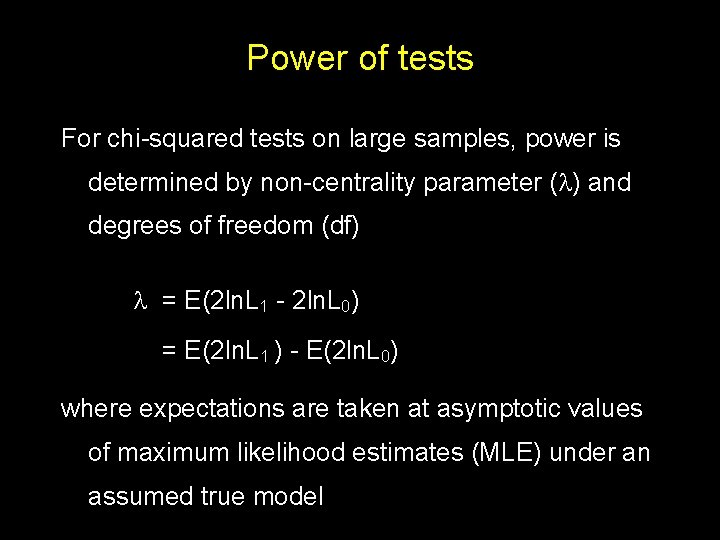
Power of tests For chi-squared tests on large samples, power is determined by non-centrality parameter ( ) and degrees of freedom (df) = E(2 ln. L 1 - 2 ln. L 0) = E(2 ln. L 1 ) - E(2 ln. L 0) where expectations are taken at asymptotic values of maximum likelihood estimates (MLE) under an assumed true model

Linkage test HA for i=j for i j H 0 for i=j for i j

Expected log likelihood under H 0 Expectation of the quadratic product is simply s, the sibship size (note: standarised trait)

Expected log likelihood under HA

Linkage test Expected NCP For sib-pairs under complete marker information Determinant of 2 -by-2 standardised covariance matrix = 1 - r 2

Approximation of NCP per sib pair is proportional to - the # of pairs in the sibship (large sibships are powerful) - the square of the additive QTL variance (decreases rapidly for QTL of v. small effect) - the sibling correlation (structure of residual variance is important)

QTL linkage POWER Type I errors Type II errors Sample N Effect Size Allele frequencies Variance explained Genetic values Marker vs functional variant Recombination fraction

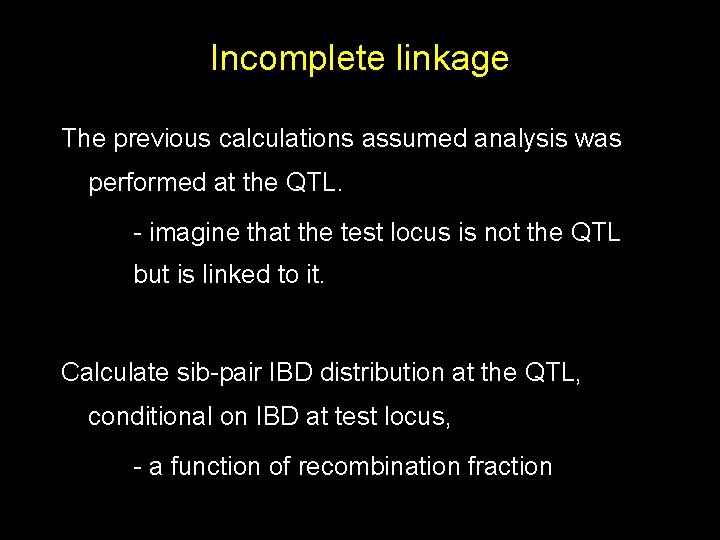
Incomplete linkage The previous calculations assumed analysis was performed at the QTL. - imagine that the test locus is not the QTL but is linked to it. Calculate sib-pair IBD distribution at the QTL, conditional on IBD at test locus, - a function of recombination fraction

at QTL at M 0 1/2 1

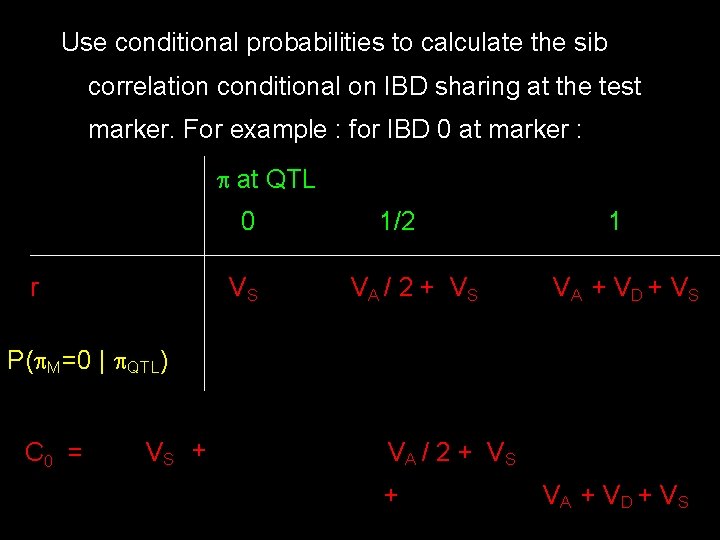
Use conditional probabilities to calculate the sib correlation conditional on IBD sharing at the test marker. For example : for IBD 0 at marker : at QTL 0 r VS 1/2 VA / 2 + V S 1 VA + V D + V S P( M=0 | QTL) C 0 = VS + VA / 2 + V S + VA + V D + V S

The noncentrality parameter per sib pair is then given by

If the QTL is additive, then attenuation of the NCP is by a factor of (1 -2 )4 = square of the correlation between the proportions of alleles IBD at two loci with recombination fraction

Effect of incomplete linkage

Effect of incomplete linkage

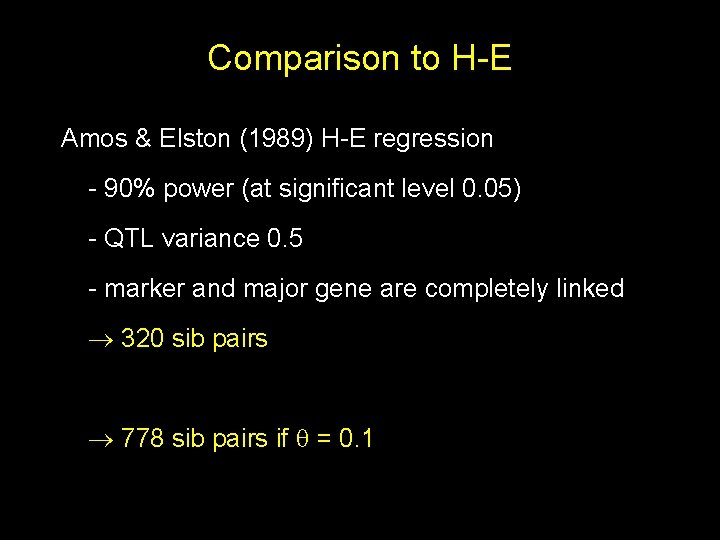
Comparison to H-E Amos & Elston (1989) H-E regression - 90% power (at significant level 0. 05) - QTL variance 0. 5 - marker and major gene are completely linked 320 sib pairs 778 sib pairs if = 0. 1

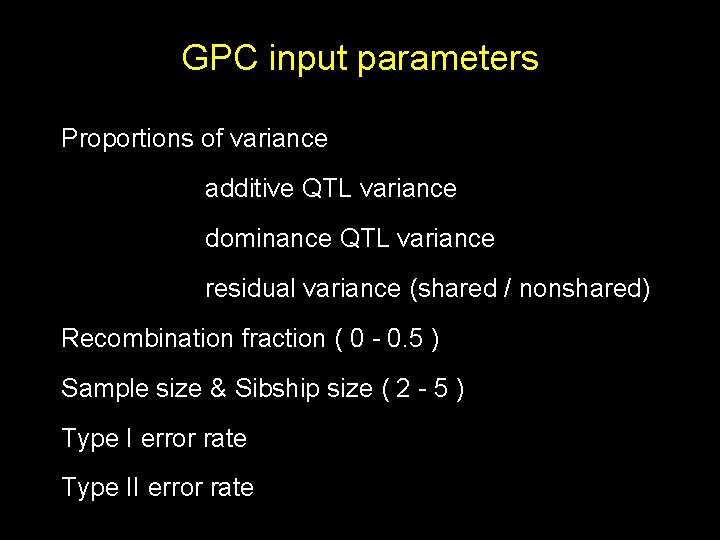
GPC input parameters Proportions of variance additive QTL variance dominance QTL variance residual variance (shared / nonshared) Recombination fraction ( 0 - 0. 5 ) Sample size & Sibship size ( 2 - 5 ) Type I error rate Type II error rate

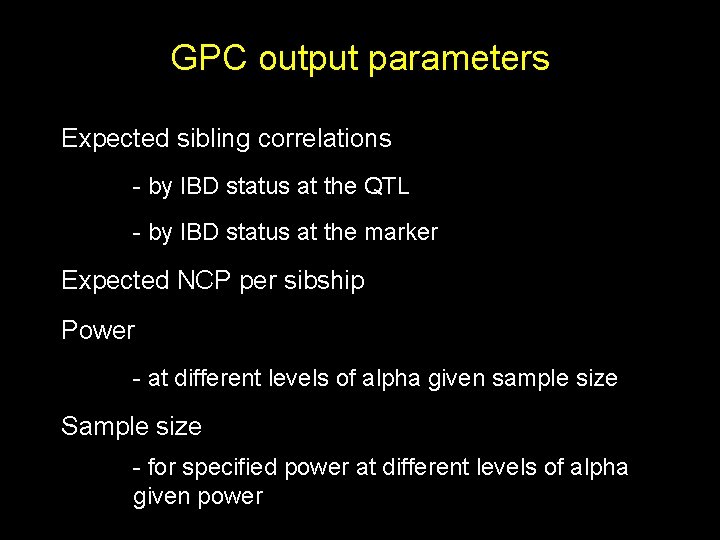
GPC output parameters Expected sibling correlations - by IBD status at the QTL - by IBD status at the marker Expected NCP per sibship Power - at different levels of alpha given sample size Sample size - for specified power at different levels of alpha given power

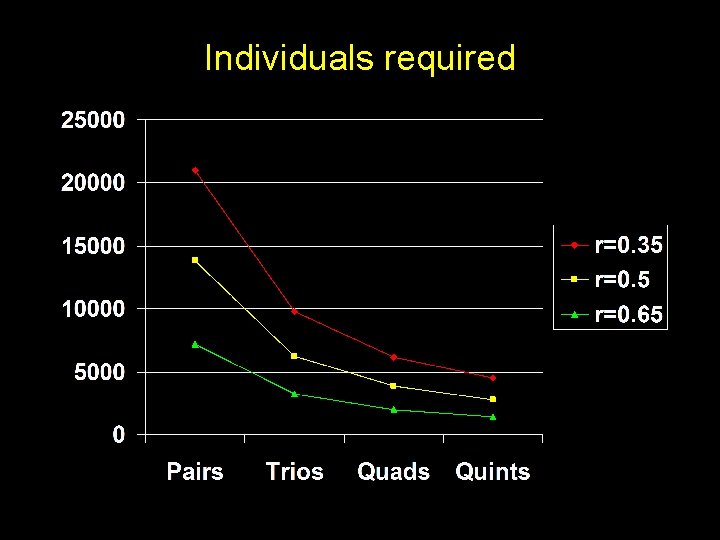
From GPC Modelling additive effects only Sibships Individuals Pairs 265 (320) 530 Pairs ( = 0. 1) 666 (778) 1332 Trios ( = 0. 1) 220 660 Quads ( = 0. 1) 110 440 Quints ( = 0. 1) 67 335

Practical Exercise 2 What is the effect on power to detect linkage of : 1. QTL variance? 2. residual sibling correlation? 3. marker-QTL recombination fraction?

Pairs required ( =0, p=0. 05, power=0. 8)

Pairs required ( =0, p=0. 05, power=0. 8)

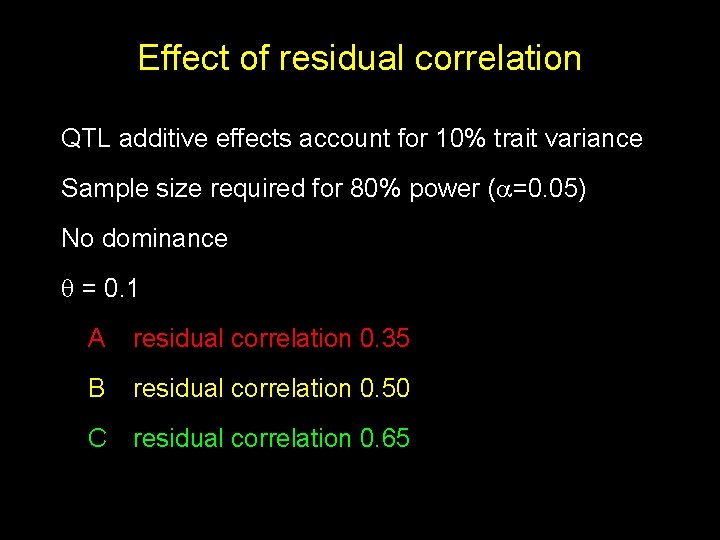
Effect of residual correlation QTL additive effects account for 10% trait variance Sample size required for 80% power ( =0. 05) No dominance = 0. 1 A residual correlation 0. 35 B residual correlation 0. 50 C residual correlation 0. 65

Individuals required

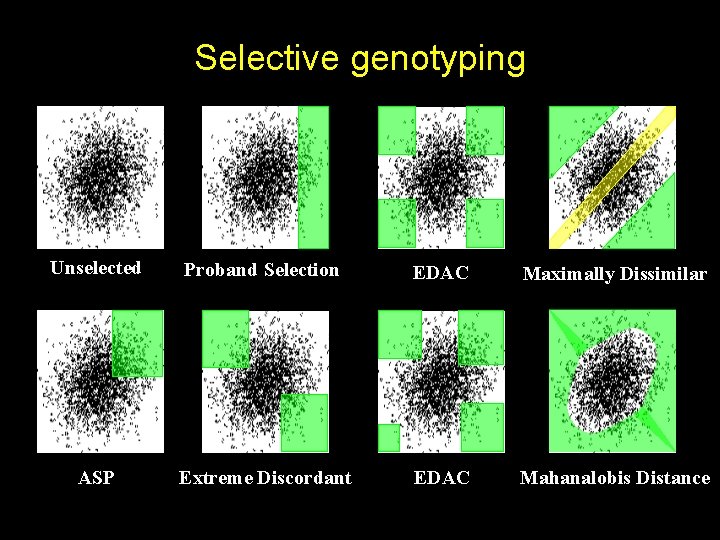
Selective genotyping Unselected Proband Selection EDAC Maximally Dissimilar ASP Extreme Discordant EDAC Mahanalobis Distance

Selective genotyping The power calculations so far assume an unselected population. - calculate expected NCP per sibship If we have a sample with trait scores - calculate expected NCP for each sibship conditional on trait values - this quantity can be used to rank order the sample for genotying

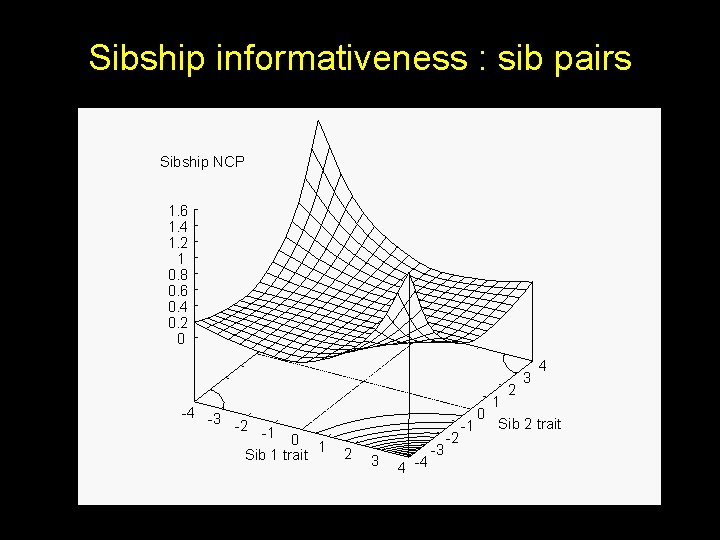
Sibship informativeness : sib pairs Sibship NCP 1. 6 1. 4 1. 2 1 0. 8 0. 6 0. 4 0. 2 0 3 -4 -3 -2 -1 0 1 Sib 1 trait 2 3 4 -4 -3 -2 4 2 1 0 Sib 2 trait -1

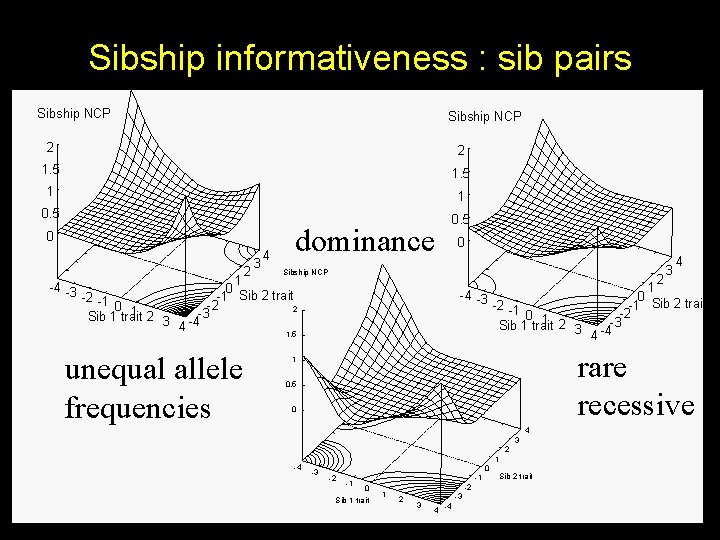
Sibship informativeness : sib pairs Sibship NCP 2 2 1. 5 1 1 0. 5 0 2 3 4 dominance 0 3 Sibship NCP 2 1 -4 -3 0 -2 -1 -1 Sib 2 trait -2 0 1 2 3 -4 -3 Sib 1 trait 4 1 -4 -3 0 -1 Sib 2 trait -2 -1 -2 0 1 2 Sib 1 trait 2 3 -4 -3 4 1. 5 unequal allele frequencies rare recessive 1 0. 5 0 4 3 2 -4 4 1 -3 0 -2 -1 -1 0 Sib 1 trait 1 -2 2 3 -3 4 -4 Sib 2 trait

Selective genotyping SEL T p d/a. 5 0 15. 82 . 1 0 17. 10 . 25 0 15. 45 . 1 1 16. 88 . 25 1 15. 76 . 5 1 18. 89 . 75 1 27. 64 . 9 43. 16 1 ASP PS ED EDAC Max. D MDis SEL B

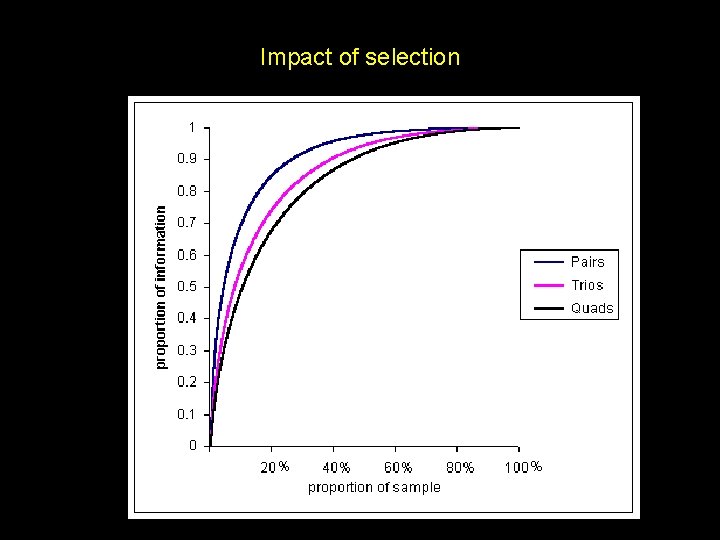
Impact of selection

QTL linkage POWER Type I errors Type II errors Sample N Effect Size Allele frequencies Variance explained Genetic values Marker vs functional variant Recombination fraction Locus informativeness PIC

Indices of marker informativeness: Markers should be highly polymorphic - alleles inherited from different sources are likely to be distinguishable Heterozygosity (H) Polymorphism Information Content (PIC) - measure number and frequency of alleles at a locus

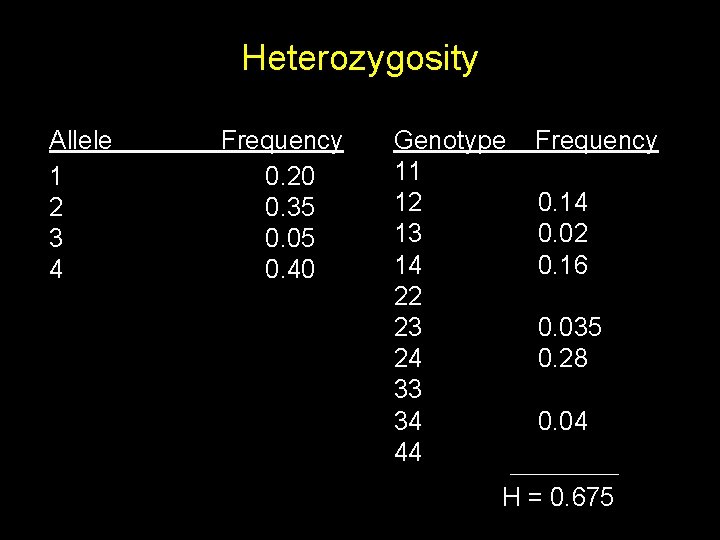
Heterozygosity n = number of alleles, pi = frequency of the ith allele. H = probability that an individual is heterozygous

Heterozygosity Allele 1 2 3 4 Frequency 0. 20 0. 35 0. 05 0. 40 Genotype 11 12 13 14 22 23 24 33 34 44 Frequency 0. 04 0. 14 0. 02 0. 16 0. 1225 0. 035 0. 28 0. 0025 0. 04 0. 16 H = 0. 675

Polymorphism information content IF a parent is heterozygous, their gametes will usually be informative. BUT if both parents & child are heterozygous for the same genotype, origins of child’s alleles are ambiguous IF C = the probability of this occurring, PIC = H - C

Polymorphism information content

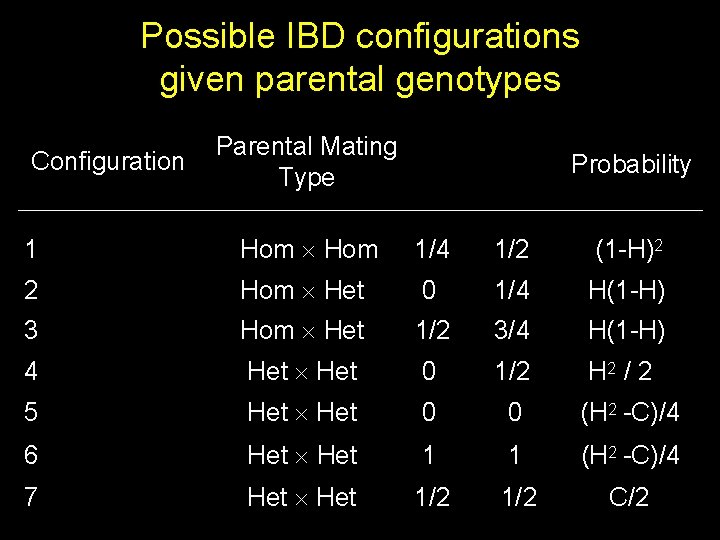
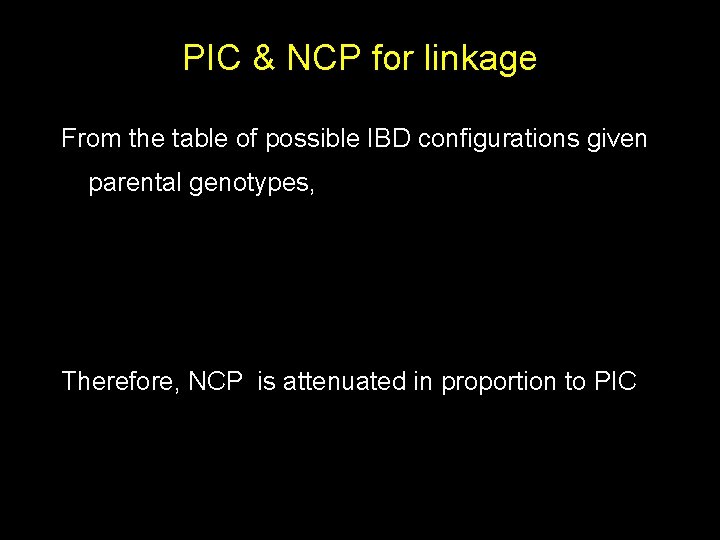
Possible IBD configurations given parental genotypes Configuration Parental Mating Type Probability 1 Hom 1/4 1/2 (1 -H)2 2 Hom Het 0 1/4 H(1 -H) 3 Hom Het 1/2 3/4 H(1 -H) 4 Het 0 1/2 H 2 / 2 5 Het 0 0 (H 2 -C)/4 6 Het 1 1 (H 2 -C)/4 7 Het 1/2 C/2

PIC & NCP for linkage From the table of possible IBD configurations given parental genotypes, Therefore, NCP is attenuated in proportion to PIC

QTL linkage POWER Type I errors Type II errors Sample N Effect Size Allele frequencies Variance explained Genetic values Marker vs functional variant Recombination fraction Locus informativeness PIC Multipoint Marker density MPIC

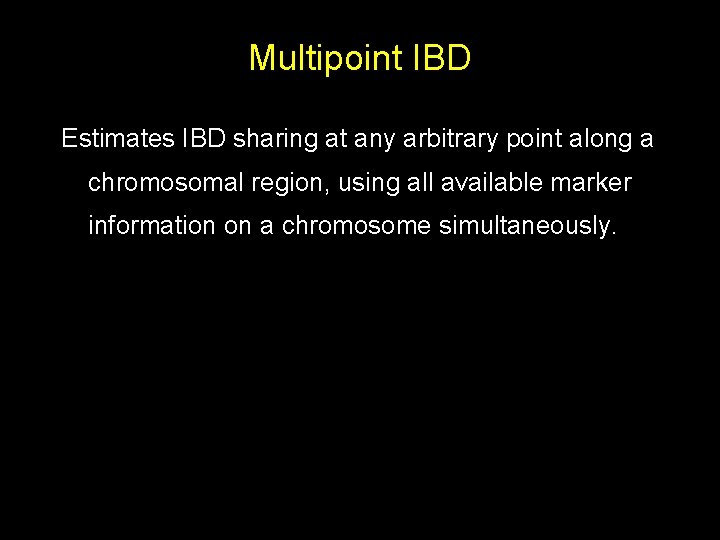
Multipoint IBD Estimates IBD sharing at any arbitrary point along a chromosomal region, using all available marker information on a chromosome simultaneously.

^ and PIC , ,

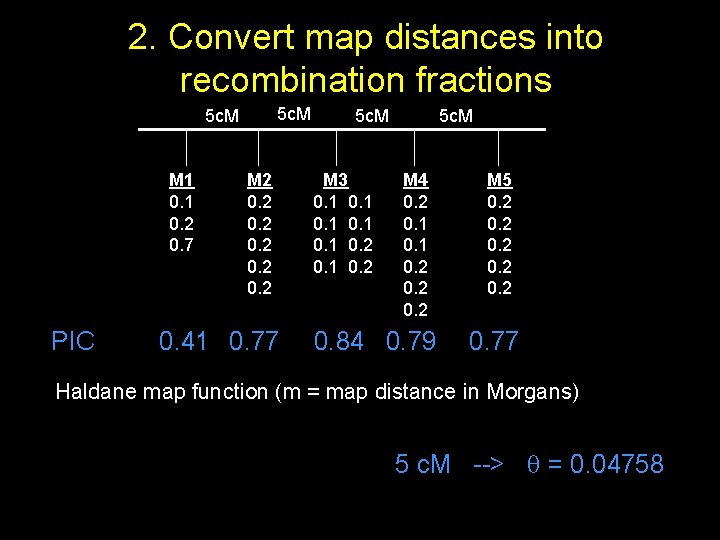
Convert PIC mapfor distances into 1. 2. Calculate each marker recombination fractions 5 c. M M 1 0. 2 0. 7 PIC M 2 0. 41 0. 77 5 c. M M 3 0. 1 0. 2 5 c. M M 4 0. 2 0. 1 0. 2 0. 84 0. 79 M 5 0. 2 0. 77 Haldane map function (m = map distance in Morgans) 5 c. M --> = 0. 04758

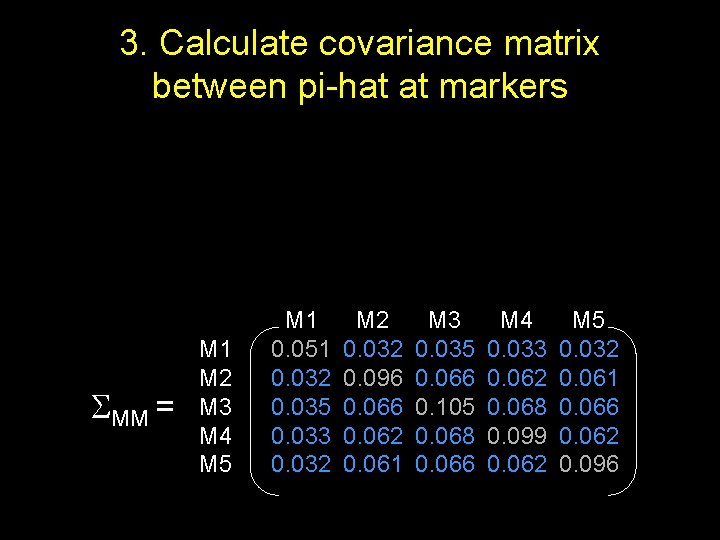
3. Calculate covariance matrix between pi-hat at markers MM = M 1 M 2 M 3 M 4 M 5 M 1 0. 051 0. 032 0. 035 0. 033 0. 032 M 2 0. 032 0. 096 0. 062 0. 061 M 3 0. 035 0. 066 0. 105 0. 068 0. 066 M 4 0. 033 0. 062 0. 068 0. 099 0. 062 M 5 0. 032 0. 061 0. 066 0. 062 0. 096

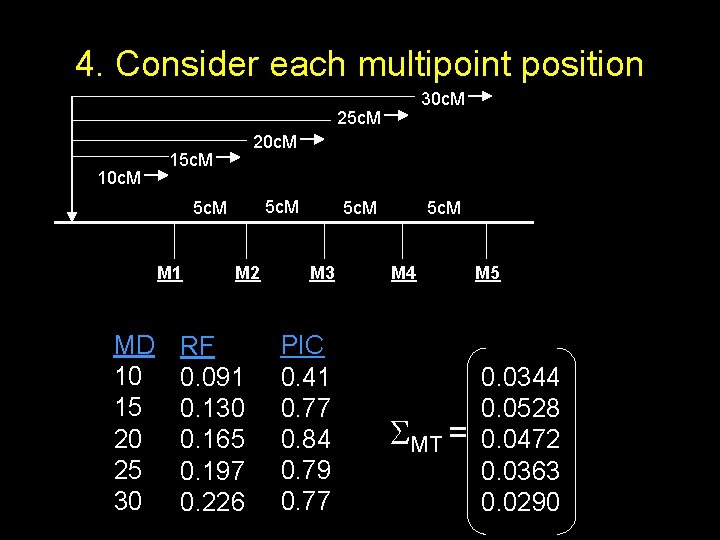
4. Consider each multipoint position 30 c. M 25 c. M At each position 20 c. M along the chromosome, calculate 15 c. M 10 c. M covariance between trait locus and each of the markers M 1 MD 10 15 20 25 30 5 c. M M 2 RF 0. 091 0. 130 0. 165 0. 197 0. 226 5 c. M M 3 PIC 0. 41 0. 77 0. 84 0. 79 0. 77 5 c. M M 4 MT = M 5 0. 0344 0. 0528 0. 0472 0. 0363 0. 0290

Fulker et al multipoint If is a vector of single marker IBD estimates then a multipoint IBD estimate at test position t is given by : Conditional on the variance of at the test position is reduced by a quantity which can be thought of as a multipoint PIC

5. Calculate MPIC

10 c. M map

5 c. M map

Exclusion mapping Exclusion : support for the hypothesis that a QTL of at least a certain effect is absent at that position Normally, the LRT compares the likelihood at the MLE and the null In exclusion mapping, the LRT compares the likelihood of a fixed effect size against the null and therefore can be negative

Conclusions Factors influencing power QTL variance Sib correlation Sibship size Marker informativeness Marker density Phenotypic selection
 Shaun purcell
Shaun purcell Linking loader and linkage editor
Linking loader and linkage editor Qtl club c'est quoi
Qtl club c'est quoi Qtl mapping
Qtl mapping Qtl mapping
Qtl mapping Qtl moodle
Qtl moodle Moonrise poem questions and answers
Moonrise poem questions and answers Victoria purcell gates
Victoria purcell gates Ralphs annual income is about $32 000
Ralphs annual income is about $32 000 Episikloid
Episikloid Stephen purcell
Stephen purcell Phil purcell planned giving
Phil purcell planned giving Zhang et al
Zhang et al Smith purcell radiation
Smith purcell radiation Smith purcell radiation
Smith purcell radiation Shaun de witt
Shaun de witt Dr michael witt
Dr michael witt Where to find pedal pulse
Where to find pedal pulse Shaun tan biography
Shaun tan biography Queen elizabeth hierarchy chart
Queen elizabeth hierarchy chart Shaun topham
Shaun topham Shaun moss
Shaun moss Shaun loke
Shaun loke Shaun harris lse
Shaun harris lse Draw the power triangle
Draw the power triangle Uht packaging
Uht packaging Pak solhin
Pak solhin Yekpan
Yekpan Pak ramlan mempunyai rumah dan kebun njop
Pak ramlan mempunyai rumah dan kebun njop Perbandingan trigonometri segitiga siku siku
Perbandingan trigonometri segitiga siku siku Angka kredit pengawas sekolah
Angka kredit pengawas sekolah Orbis tetrapak
Orbis tetrapak Elemen kemahiran abad ke 21
Elemen kemahiran abad ke 21 Pak amin membeli beras di pasar termasuk kegiatan
Pak amin membeli beras di pasar termasuk kegiatan Tetra pak ii case
Tetra pak ii case Hypermetropi
Hypermetropi Trilles maureilhan
Trilles maureilhan Tehno eko pak
Tehno eko pak Dari hasil tes ternyata urin pak yudha mengandung glukosa
Dari hasil tes ternyata urin pak yudha mengandung glukosa Logo kelompok 1
Logo kelompok 1 Keuntungan yang diperoleh pak karta semakin bertambah
Keuntungan yang diperoleh pak karta semakin bertambah Pak life
Pak life Sebuah perusahaan percetakan memberikan rabat 30
Sebuah perusahaan percetakan memberikan rabat 30 Zweet pak
Zweet pak Unsur utama dan unsur penunjang skp guru
Unsur utama dan unsur penunjang skp guru Hal yang memicu penghentian pendarahan darah adalah
Hal yang memicu penghentian pendarahan darah adalah Renungan ulangan 6 4-9
Renungan ulangan 6 4-9 Rumus evaluasi pembelajaran
Rumus evaluasi pembelajaran Pak naw
Pak naw Pak bowo pengrajin wayang golek
Pak bowo pengrajin wayang golek Robert pak
Robert pak Access quarterly packages
Access quarterly packages Kampuchea
Kampuchea Bastian berusia 3 tahun lebih tua dari diah
Bastian berusia 3 tahun lebih tua dari diah Pak vsceng
Pak vsceng Sella turcica
Sella turcica Umur pak agus 3 kali umur iwan
Umur pak agus 3 kali umur iwan Pak sham
Pak sham Diya pakistan status
Diya pakistan status Kata nama khas
Kata nama khas Anu pak
Anu pak Linkage institutions definition
Linkage institutions definition Purpose of steering system
Purpose of steering system Ackerman linkage
Ackerman linkage Punnet square of hemophilia
Punnet square of hemophilia X linked dominant inheritance punnett square
X linked dominant inheritance punnett square Sex rat
Sex rat Section 3 gene linkage and polyploidy
Section 3 gene linkage and polyploidy Nrlm,bank linkage portal login
Nrlm,bank linkage portal login Evidence linkage triangle
Evidence linkage triangle Graphical linkage synthesis
Graphical linkage synthesis Media linkage institution
Media linkage institution