Relationship between quantitative trait inheritance and genetic markers

Relationship between quantitative trait inheritance and genetic markers ( A rationale for QTL mapping) (1) Genes controlling quantitative traits are located in the genome (2) just like simple genetic markers. (2) If markers cover a large proportion of the genome then there is a chance that some genes controlling quantitative traits are linked with the some of these markers. (3) If genes and markers are segregating in a genetically defined population, then the linkage relationship may be resolved by studying the association between trait variation and marker segregation pattern.
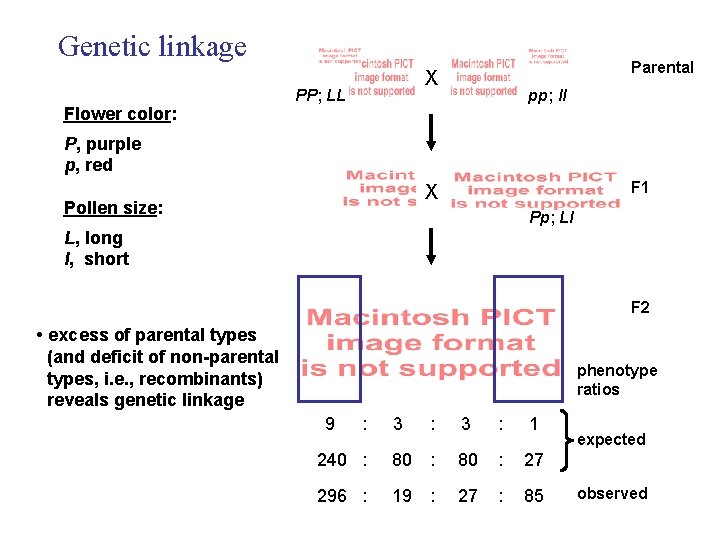
Genetic linkage Parental X PP; LL pp; ll Flower color: P, purple p, red F 1 X Pollen size: Pp; Ll L, long l, short F 2 • excess of parental types (and deficit of non-parental types, i. e. , recombinants) reveals genetic linkage phenotype ratios 9 : 3 : 1 240 : 80 : 27 296 : 19 : 27 : 85 expected observed
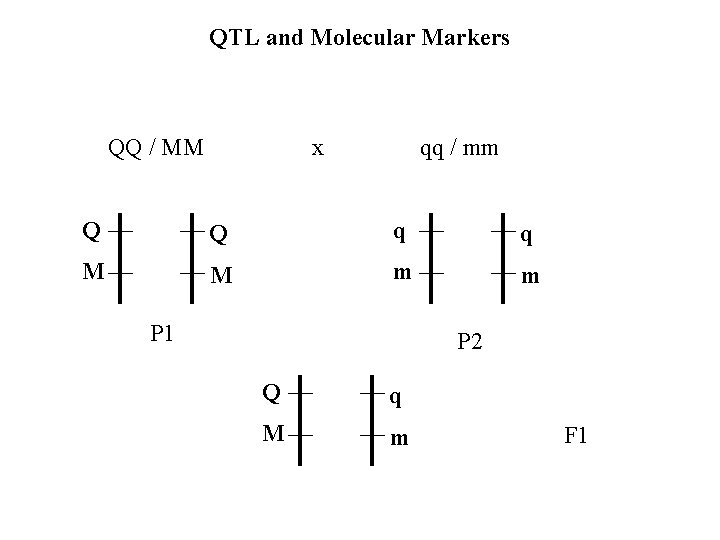
QTL and Molecular Markers QQ / MM x qq / mm Q Q q q M M m m P 1 P 2 Q q M m F 1
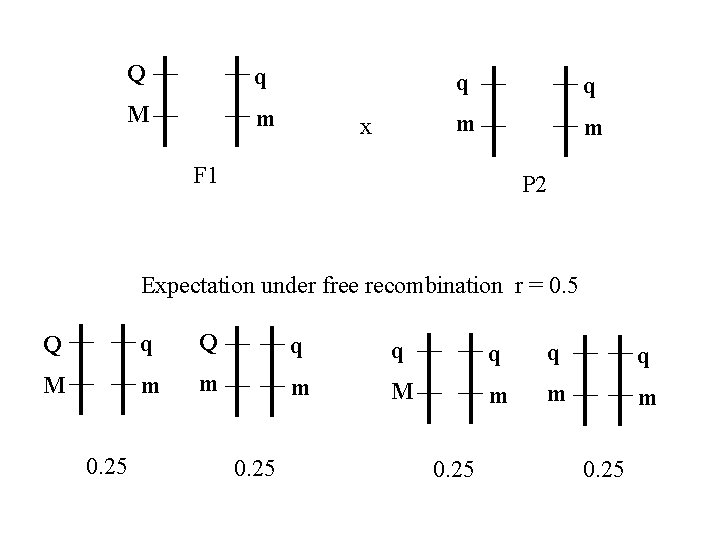
Q q M m x q q m m F 1 P 2 Expectation under free recombination r = 0. 5 Q q q q M m m m 0. 25
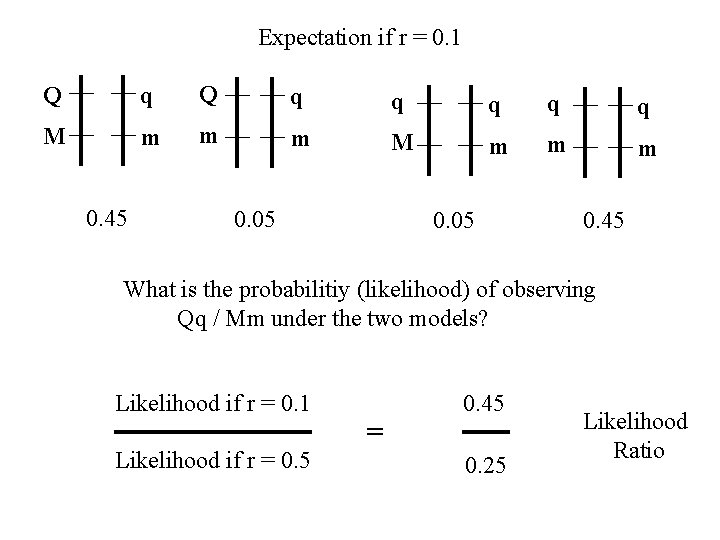
Expectation if r = 0. 1 Q q q q M m m m 0. 45 0. 05 0. 45 What is the probabilitiy (likelihood) of observing Qq / Mm under the two models? Likelihood if r = 0. 1 Likelihood if r = 0. 5 = 0. 45 0. 25 Likelihood Ratio
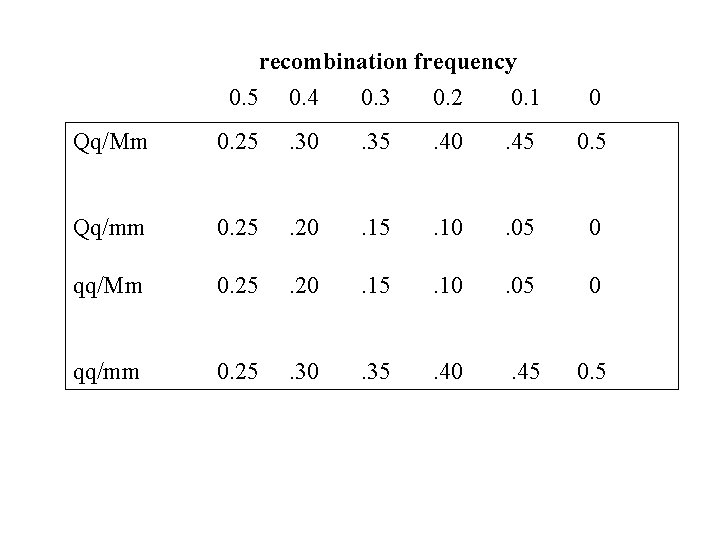
recombination frequency 0. 5 0. 4 0. 3 0. 2 0. 1 0 Qq/Mm 0. 25 . 30 . 35 . 40 . 45 0. 5 Qq/mm 0. 25 . 20 . 15 . 10 . 05 0 qq/Mm 0. 25 . 20 . 15 . 10 . 05 0 qq/mm 0. 25 . 30 . 35 . 40 . 45 0. 5
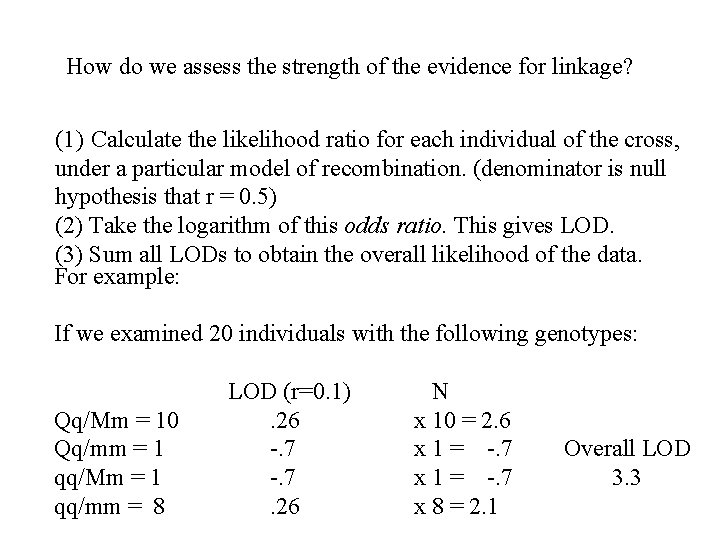
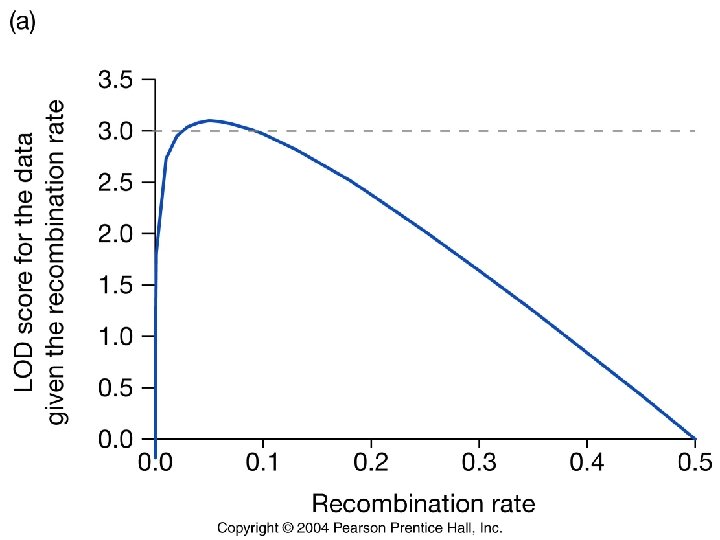
How do we assess the strength of the evidence for linkage? (1) Calculate the likelihood ratio for each individual of the cross, under a particular model of recombination. (denominator is null hypothesis that r = 0. 5) (2) Take the logarithm of this odds ratio. This gives LOD. (3) Sum all LODs to obtain the overall likelihood of the data. For example: If we examined 20 individuals with the following genotypes: Qq/Mm = 10 Qq/mm = 1 qq/Mm = 1 qq/mm = 8 LOD (r=0. 1). 26 -. 7. 26 N x 10 = 2. 6 x 1 = -. 7 x 8 = 2. 1 Overall LOD 3. 3
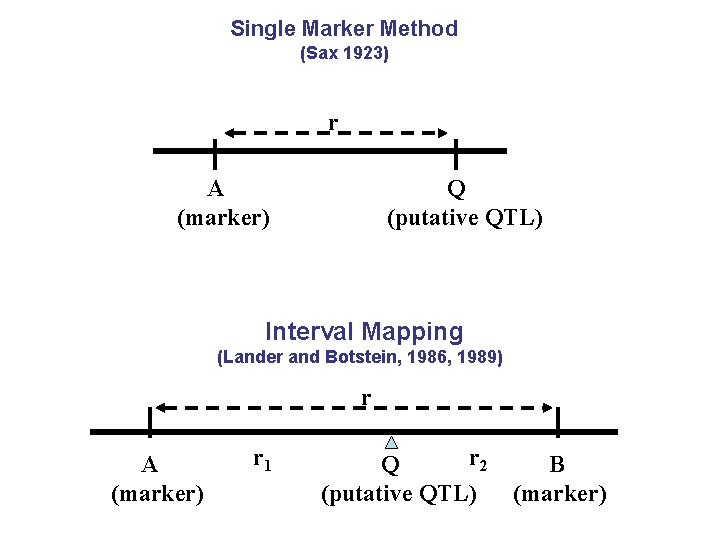
Single Marker Method (Sax 1923) r A (marker) Q (putative QTL) Interval Mapping (Lander and Botstein, 1986, 1989) r A (marker) r 1 r 2 Q (putative QTL) B (marker)
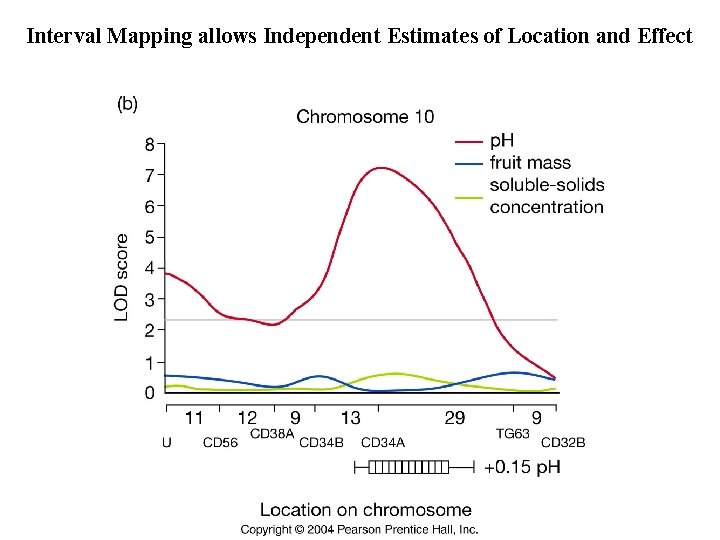
Interval Mapping allows Independent Estimates of Location and Effect

QTL affecting tomato size or shape
How Many Genetic Differences Underlie Evolutionarily Important Traits?
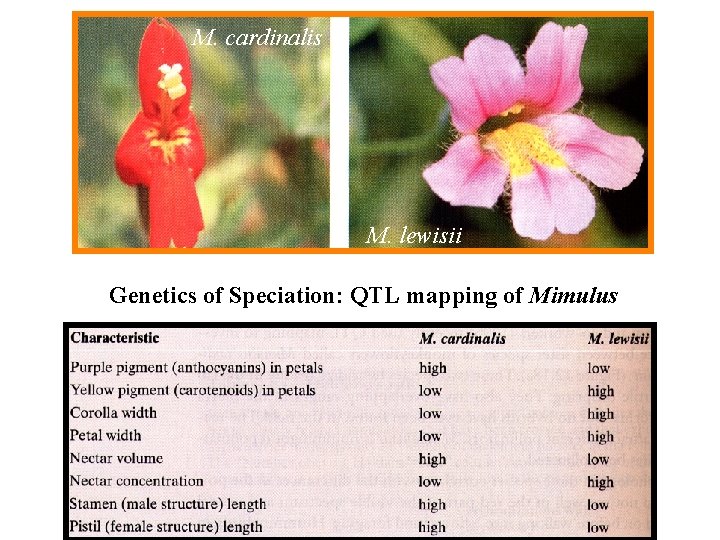
M. cardinalis M. lewisii Genetics of Speciation: QTL mapping of Mimulus
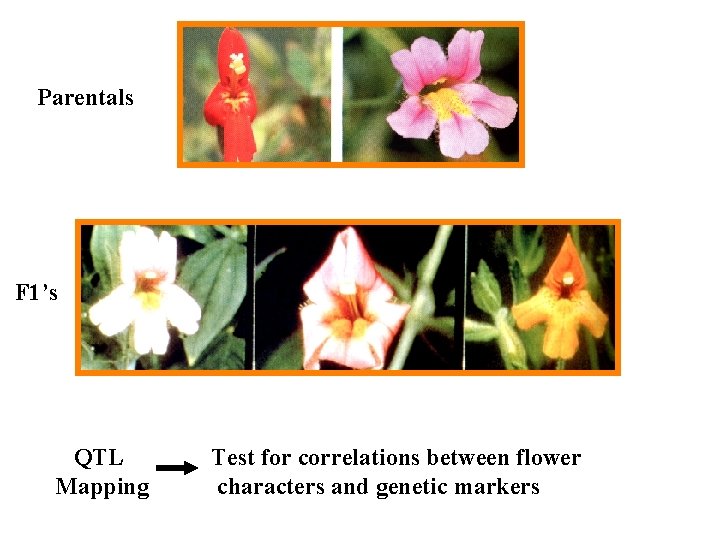
Parentals F 1’s QTL Mapping Test for correlations between flower characters and genetic markers
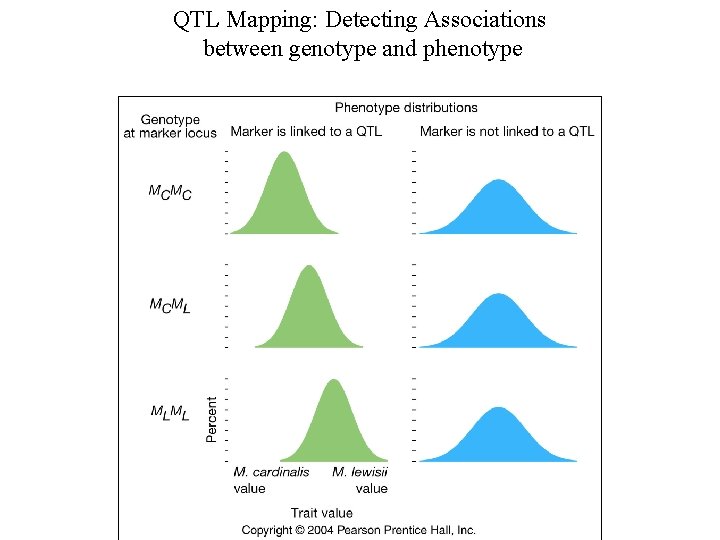
QTL Mapping: Detecting Associations between genotype and phenotype
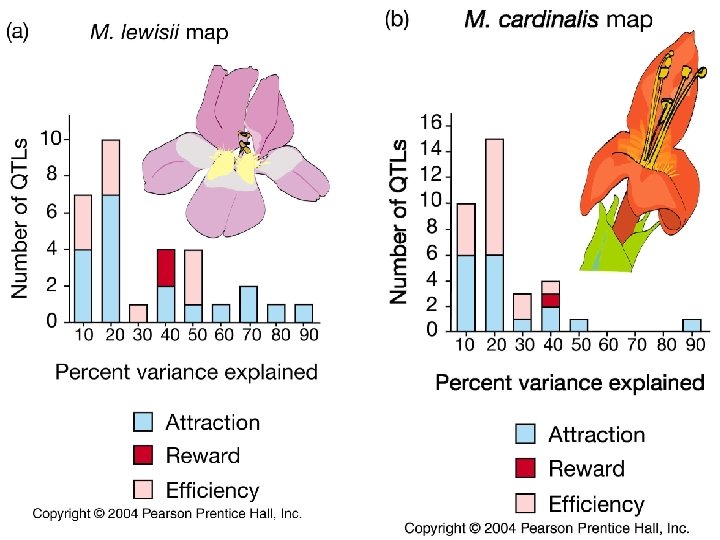
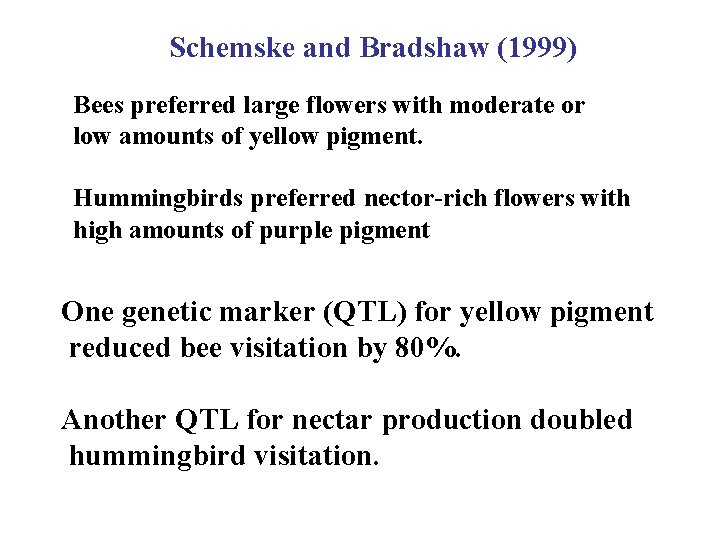
Schemske and Bradshaw (1999) Bees preferred large flowers with moderate or low amounts of yellow pigment. Hummingbirds preferred nector-rich flowers with high amounts of purple pigment One genetic marker (QTL) for yellow pigment reduced bee visitation by 80%. Another QTL for nectar production doubled hummingbird visitation.
- Slides: 17