Epistasis Multilocus Modelling Shaun Purcell Pak Sham SGDP









- Slides: 37

Epistasis / Multi-locus Modelling Shaun Purcell, Pak Sham SGDP, Io. P, London, UK

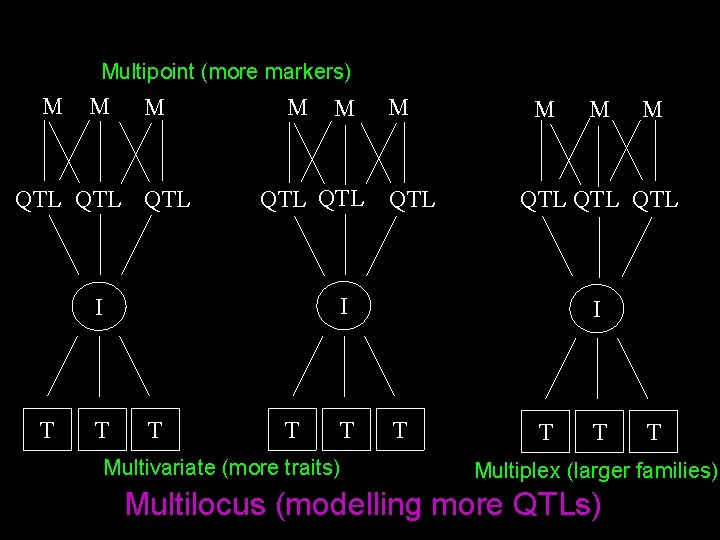
Multipoint (more markers) M M M QTL QTL M T T Multivariate (more traits) M M QTL QTL I I T M I T T Multiplex (larger families) Multilocus (modelling more QTLs)

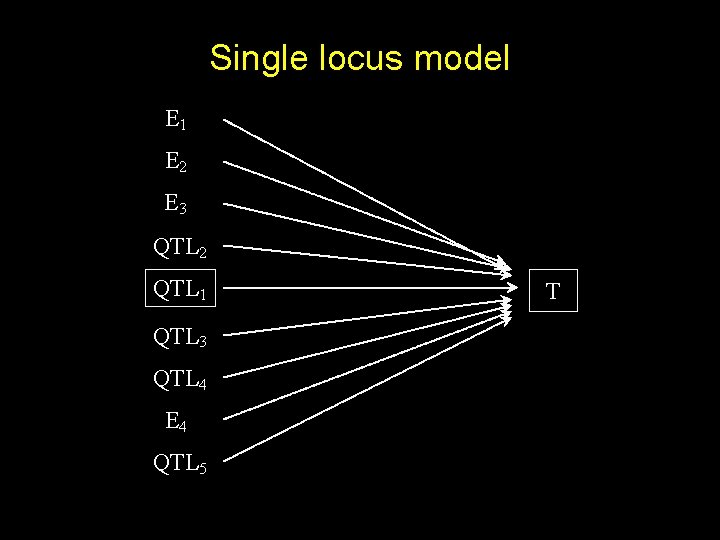
Single locus model E 1 E 2 E 3 QTL 2 QTL 1 QTL 3 QTL 4 E 4 QTL 5 T

Multilocus model E 1 E 2 E 3 QTL 1 QTL 2 QTL 4 E 4 QTL 5 T

GENE x GENE Interaction GENE x GENE INTERACTION : Epistasis Additive genetic effects : alleles at a locus and across loci independently sum to result in a net phenotypic effect Nonadditive genetic effects : effects of an allele modified by the presence of other alleles (either at the same locus or at different loci)

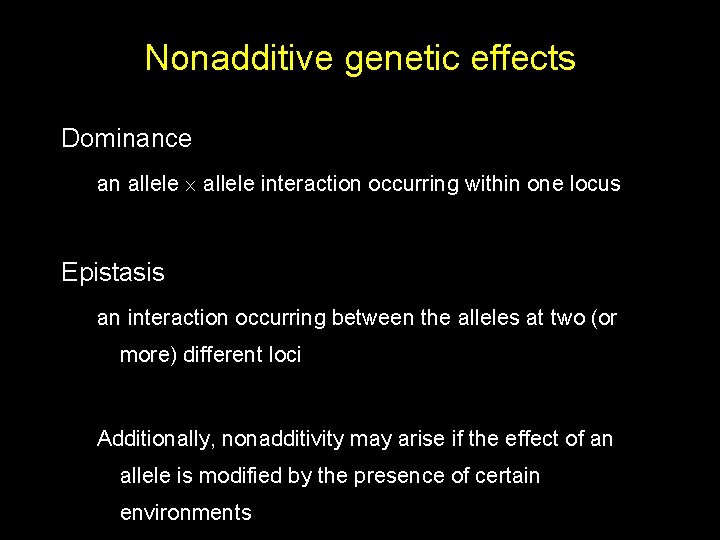
Nonadditive genetic effects Dominance an allele interaction occurring within one locus Epistasis an interaction occurring between the alleles at two (or more) different loci Additionally, nonadditivity may arise if the effect of an allele is modified by the presence of certain environments

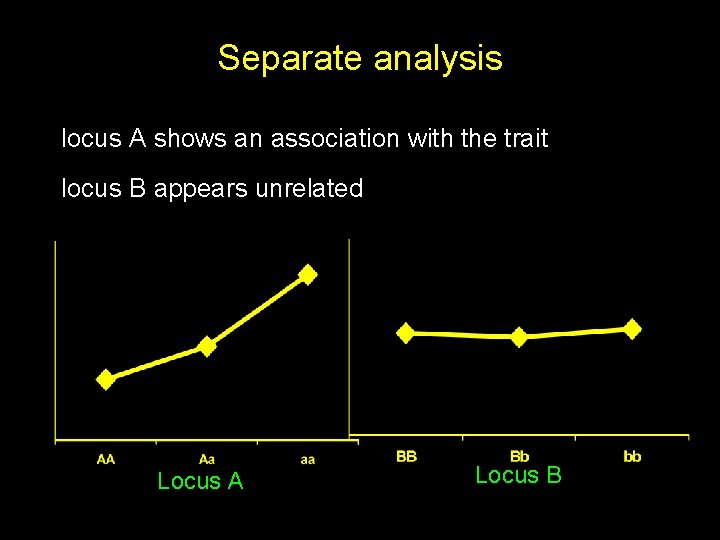
Separate analysis locus A shows an association with the trait locus B appears unrelated Locus A Locus B

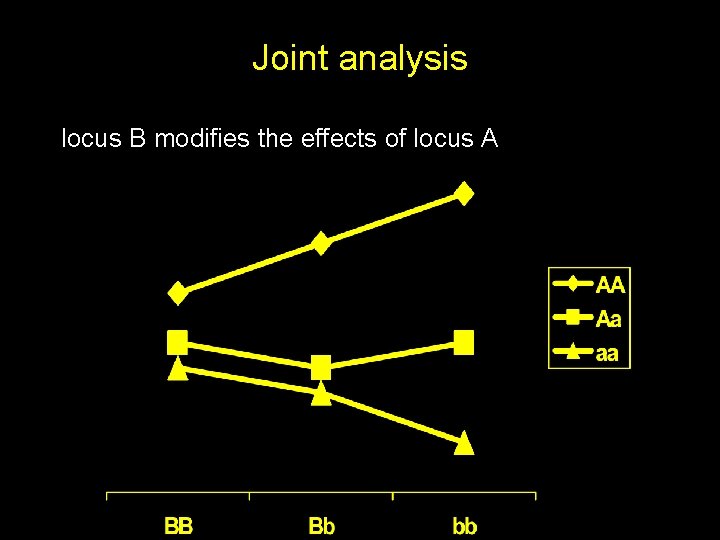
Joint analysis locus B modifies the effects of locus A

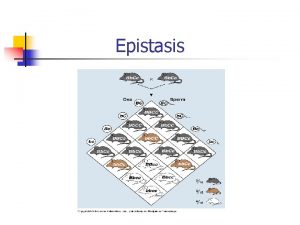
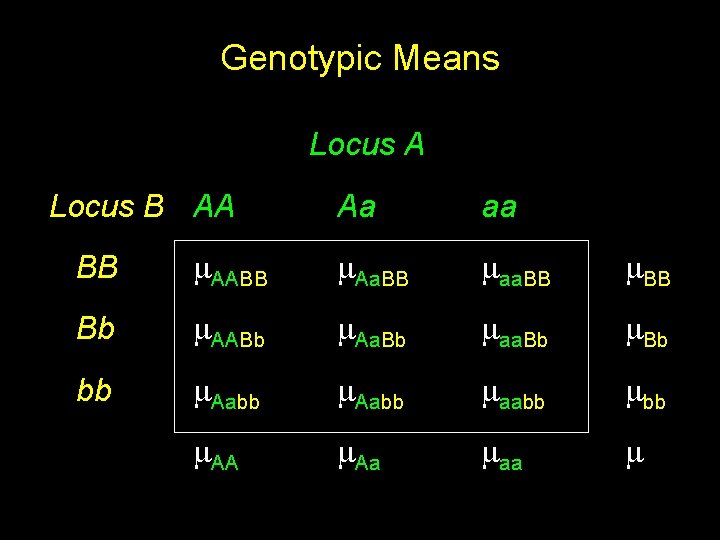
Genotypic Means Locus A Locus B AA Aa aa BB AABB Aa. BB aa. BB Bb AABb Aa. Bb aa. Bb bb Aabb aabb AA Aa aa

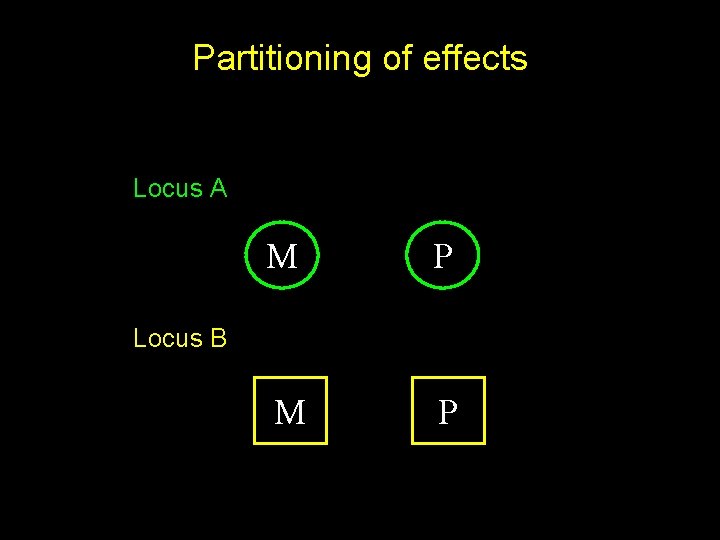
Partitioning of effects Locus A M P Locus B

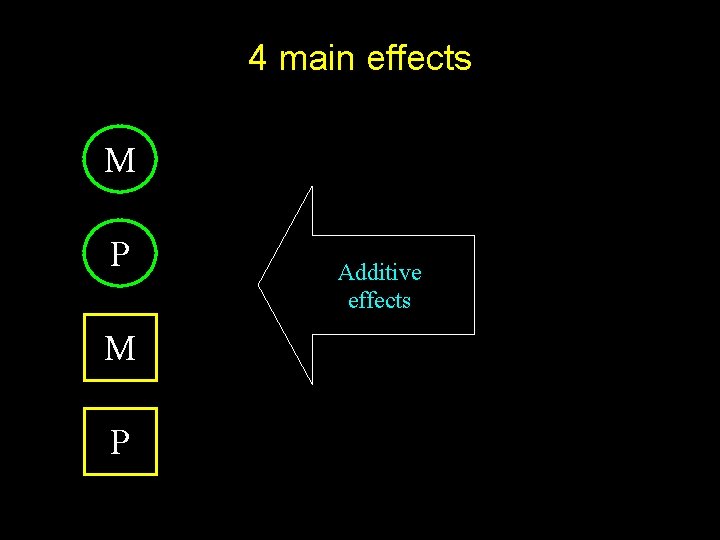
4 main effects M P Additive effects

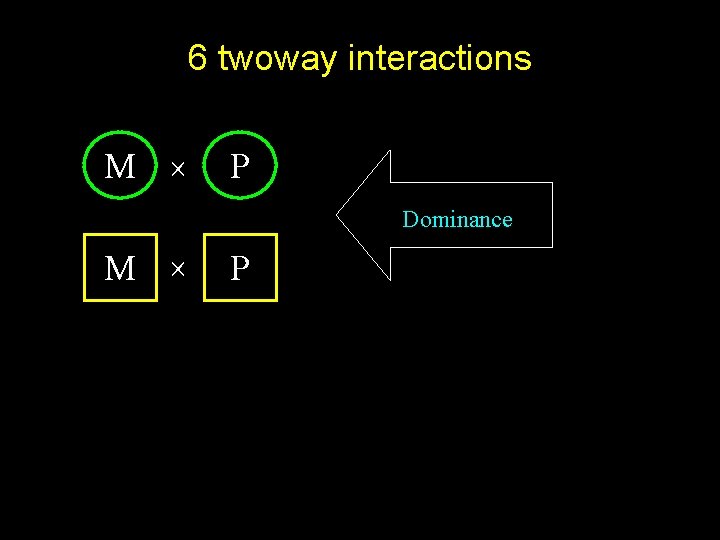
6 twoway interactions M P Dominance M P

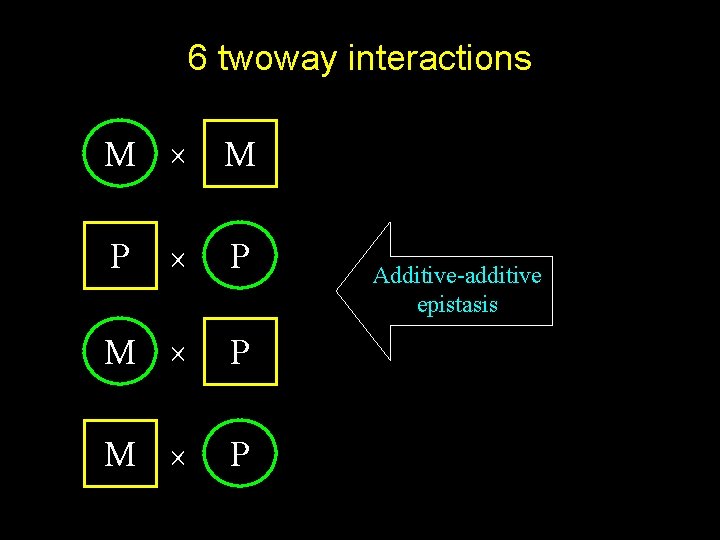
6 twoway interactions M M P M P P Additive-additive epistasis

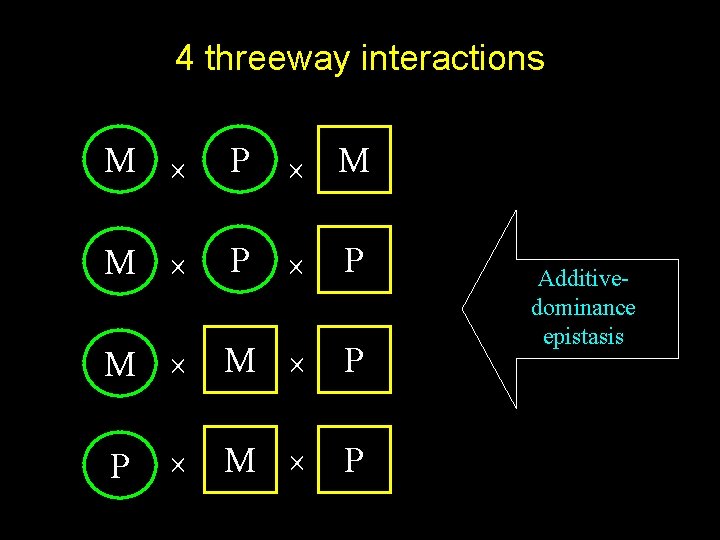
4 threeway interactions M P M M P P M M P P Additivedominance epistasis

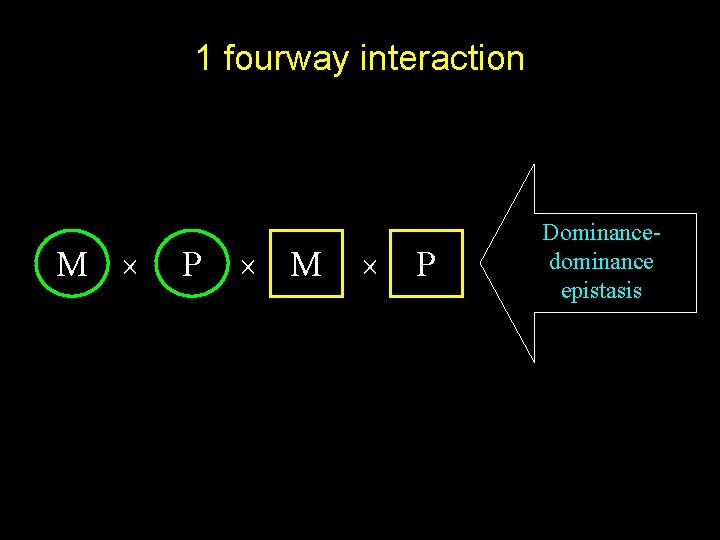
1 fourway interaction M P M P Dominancedominance epistasis

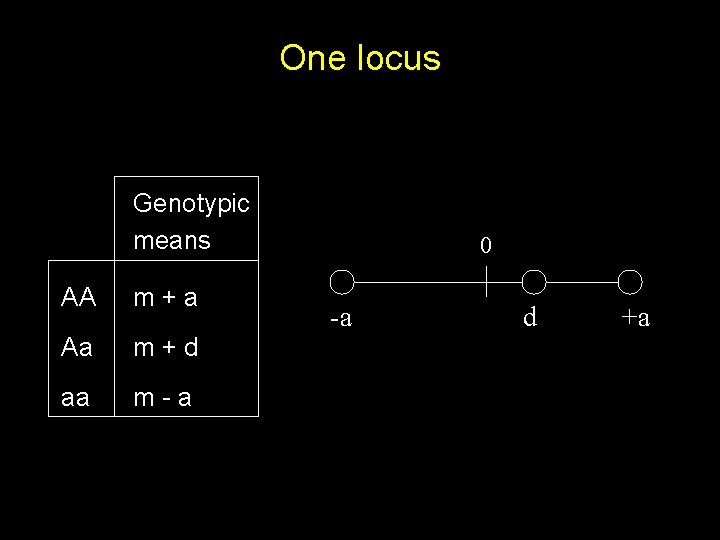
One locus Genotypic means AA m+a Aa m+d aa m-a 0 -a d +a

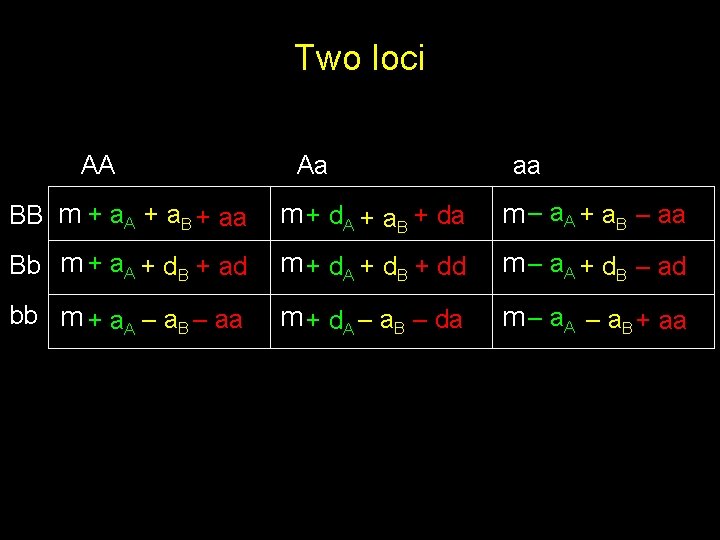
Two loci AA Aa aa BB m + a. A + a. B + aa m + d. A + a. B + da m – a. A + a. B – aa Bb m + a. A + d. B + ad m + d. A + d. B + dd m – a. A + d. B – ad bb m + a. A – a. B – aa m + d. A – a. B – da m – a. A – a. B + aa

Research questions How can epistasis be modelled under a variance components framework? How powerful is QTL linkage to detect epistasis? How does the presence of epistasis impact QTL detection when epistasis is not modelled?

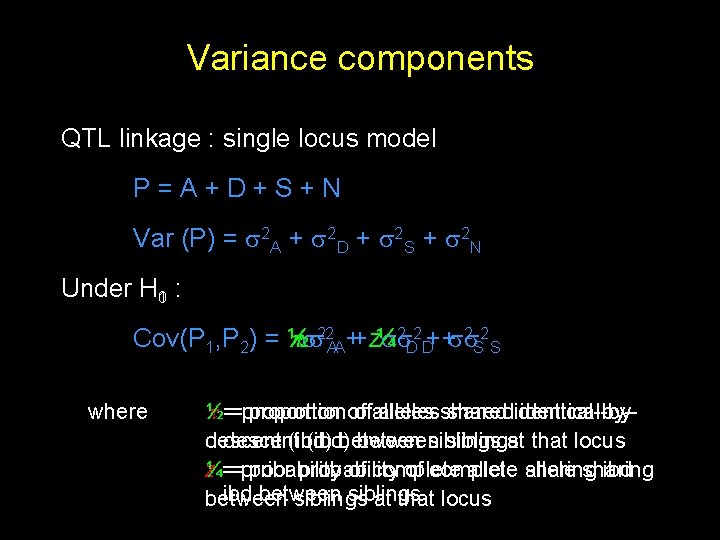
Variance components QTL linkage : single locus model P=A+D+S+N Var (P) = 2 A + 2 D + 2 S + 2 N Under H 01 : 2 2 ++ 2 2 Cov(P 1, P 2) = ½ 22 AA++z ¼ DD SS where ½==proportion proportionof ofallelessharedidentical-bydescent (ibd) between siblings at that locus z ==probability ¼ prior probability of complete allele sharing ibd between siblings at that locus

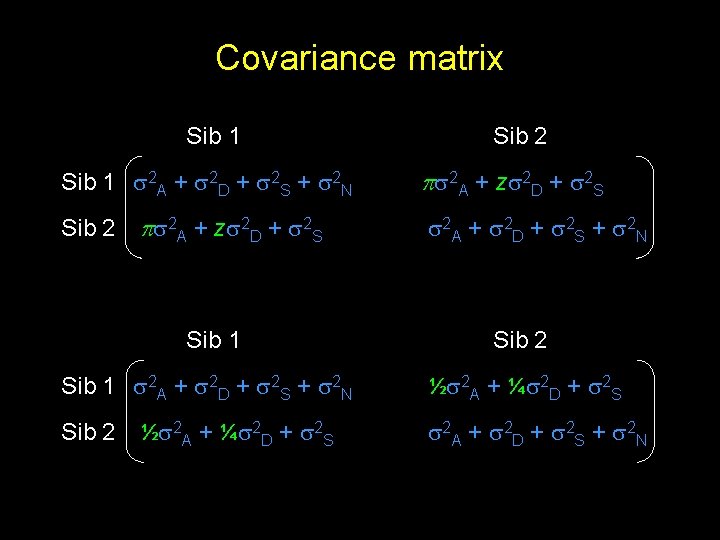
Covariance matrix Sib 1 2 A + 2 D + 2 S + 2 N Sib 2 2 A + z 2 D + 2 S Sib 1 2 A + 2 D + 2 S + 2 N Sib 2 ½ 2 A + ¼ 2 D + 2 S Sib 2 2 A + z 2 D + 2 S 2 A + 2 D + 2 S + 2 N Sib 2 ½ 2 A + ¼ 2 D + 2 S 2 A + 2 D + 2 S + 2 N

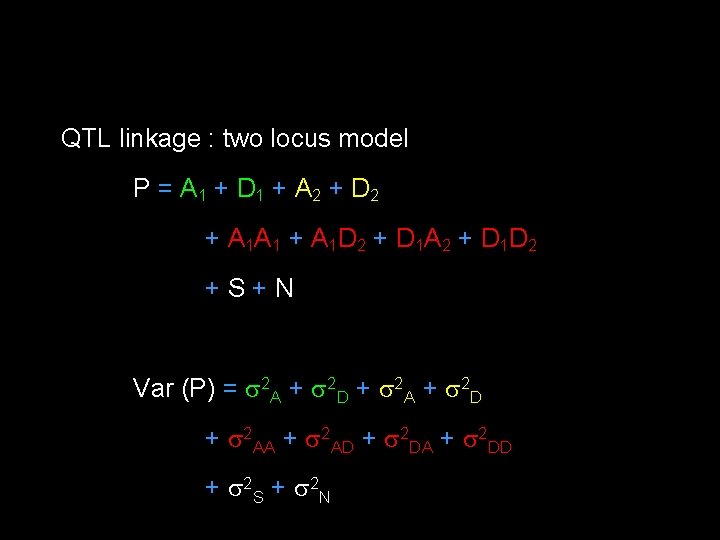
QTL linkage : two locus model P = A 1 + D 1 + A 2 + D 2 + A 1 A 1 + A 1 D 2 + D 1 A 2 + D 1 D 2 +S+N Var (P) = 2 A + 2 D + 2 AA + 2 AD + 2 DA + 2 DD + 2 S + 2 N

Under linkage : Cov(P 1, P 2) = 2 A + z 2 D + 2 A + z 2 AD + z 2 DA + zz 2 DD + 2 S Under null : Cov(P 1, P 2) = ½ 2 A + ¼ 2 D + E( ) 2 A+E( z) 2 AD +E(z ) 2 DA+ E(zz) 2 DD + 2 S

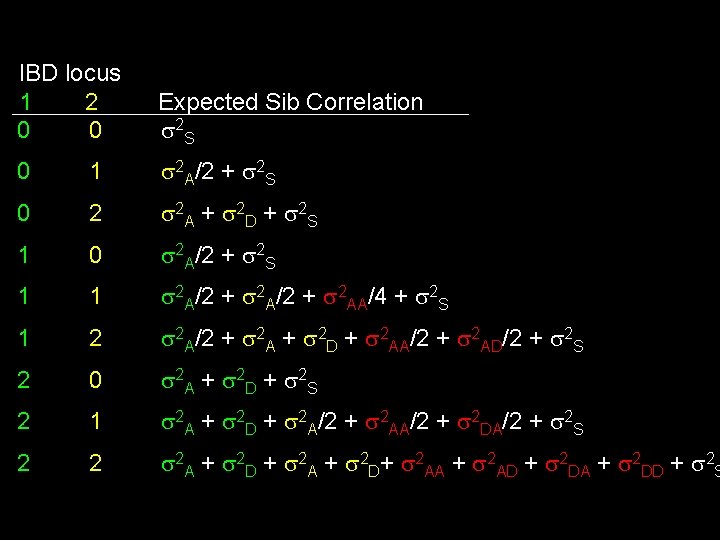
IBD locus 1 2 0 0 Expected Sib Correlation 2 S 0 1 2 A/2 + 2 S 0 2 2 A + 2 D + 2 S 1 0 2 A/2 + 2 S 1 1 2 A/2 + 2 AA/4 + 2 S 1 2 2 A/2 + 2 A + 2 D + 2 AA/2 + 2 AD/2 + 2 S 2 0 2 A + 2 D + 2 S 2 1 2 A + 2 D + 2 A/2 + 2 AA/2 + 2 DA/2 + 2 S 2 2 2 A + 2 D + 2 A + 2 D+ 2 AA + 2 AD + 2 DA + 2 DD + 2 S

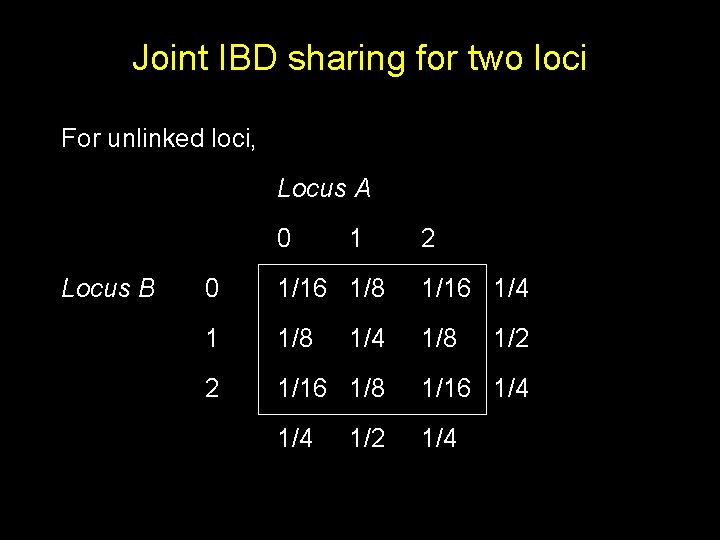
Joint IBD sharing for two loci For unlinked loci, Locus A 0 Locus B 1 2 0 1/16 1/8 1/16 1/4 1 1/8 2 1/16 1/8 1/16 1/4 1/4 1/2

Joint IBD sharing for two linked loci at QTL 2 0 1/2 1 at QTL 1 0 1/2 1

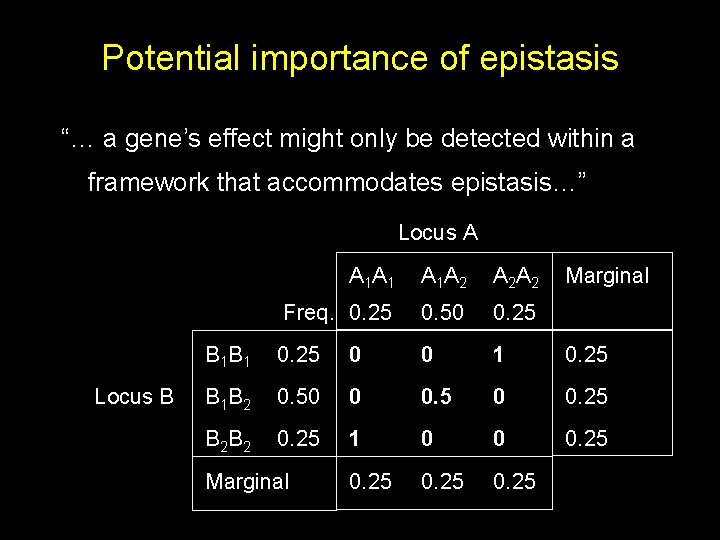
Potential importance of epistasis “… a gene’s effect might only be detected within a framework that accommodates epistasis…” Locus A Locus B A 1 A 1 A 1 A 2 A 2 A 2 Freq. 0. 25 0. 50 0. 25 B 1 B 1 0. 25 0 0 1 0. 25 B 1 B 2 0. 50 0 0. 5 0 0. 25 B 2 B 2 0. 25 1 0 0 0. 25 Marginal

Power calculations for epistasis Specify genotypic means, allele frequencies residual variance Calculate under full model and submodels variance components expected non-centrality parameter (NCP)

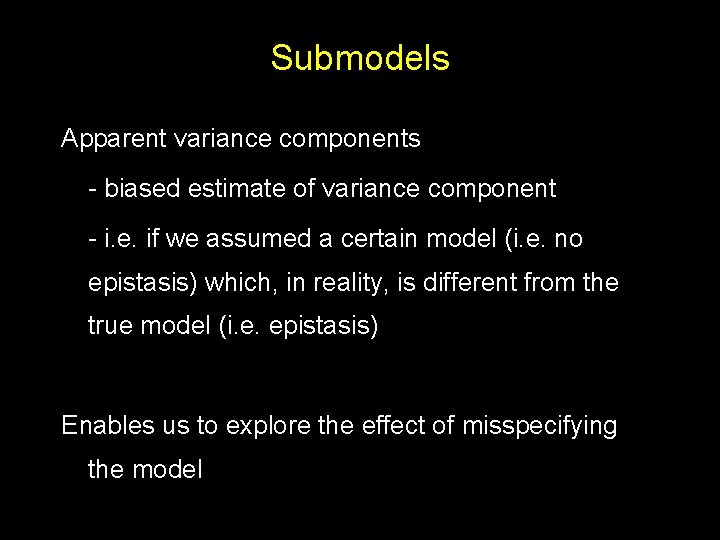
Submodels Apparent variance components - biased estimate of variance component - i. e. if we assumed a certain model (i. e. no epistasis) which, in reality, is different from the true model (i. e. epistasis) Enables us to explore the effect of misspecifying the model

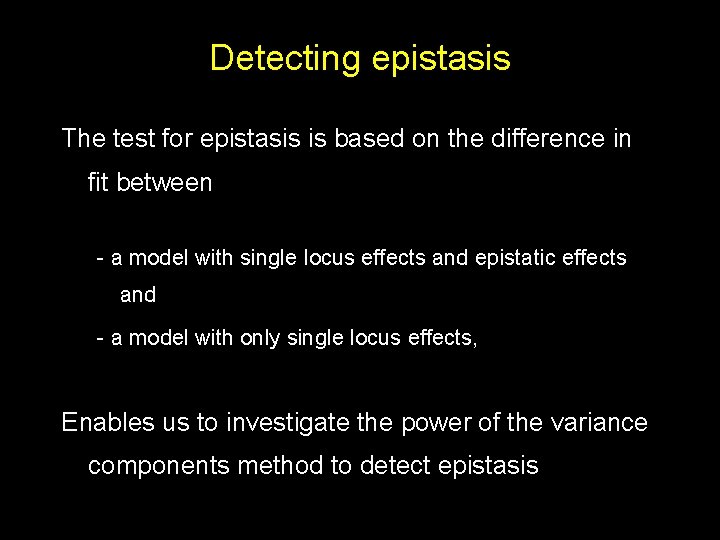
Detecting epistasis The test for epistasis is based on the difference in fit between - a model with single locus effects and epistatic effects and - a model with only single locus effects, Enables us to investigate the power of the variance components method to detect epistasis

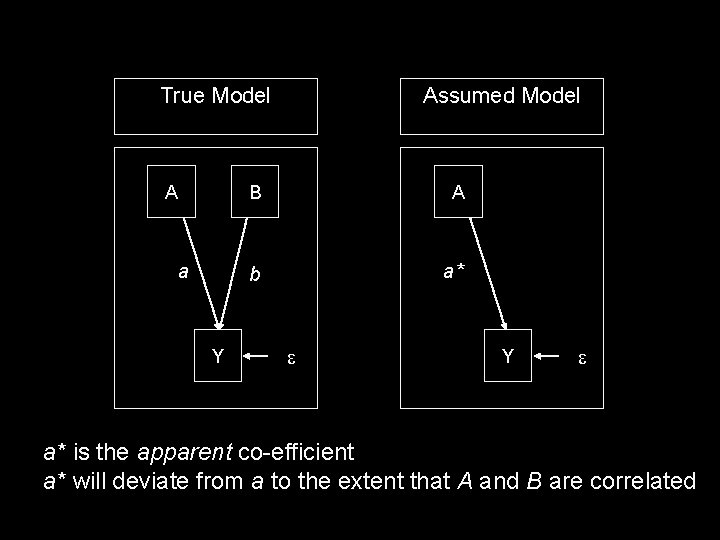
True Model A a Y Assumed Model B A b a* Y a* is the apparent co-efficient a* will deviate from a to the extent that A and B are correlated

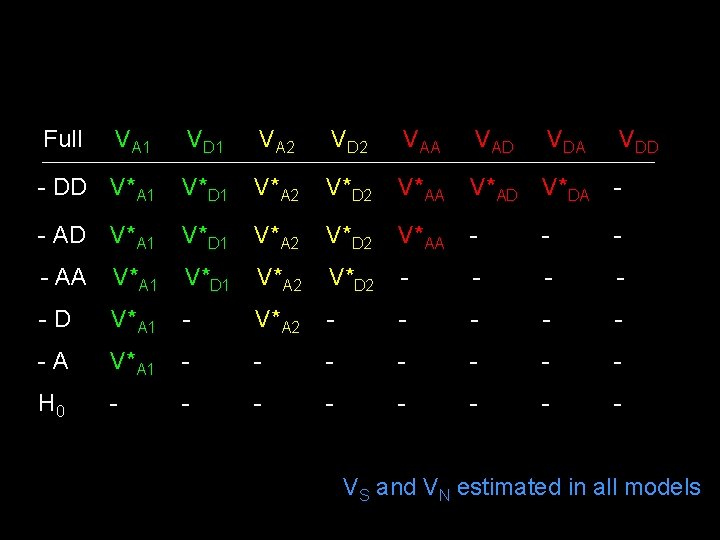
Full VA 1 VD 1 VA 2 VD 2 VAA VAD VDA VDD - DD V*A 1 V*D 1 V*A 2 V*D 2 V*AA V*AD V*DA - - AD V*A 1 V*D 1 V*A 2 V*D 2 V*AA - - AA V*A 1 V*D 1 V*A 2 V*D 2 - - -D V*A 1 - V*A 2 - - -A V*A 1 - - - - H 0 - - - - VS and VN estimated in all models

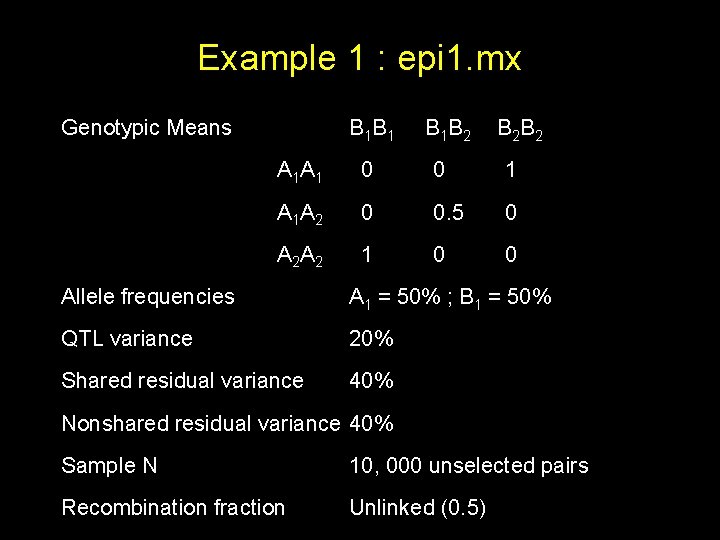
Example 1 : epi 1. mx Genotypic Means B 1 B 1 B 1 B 2 B 2 B 2 A 1 A 1 0 0 1 A 1 A 2 0 0. 5 0 A 2 A 2 1 0 0 Allele frequencies A 1 = 50% ; B 1 = 50% QTL variance 20% Shared residual variance 40% Nonshared residual variance 40% Sample N 10, 000 unselected pairs Recombination fraction Unlinked (0. 5)

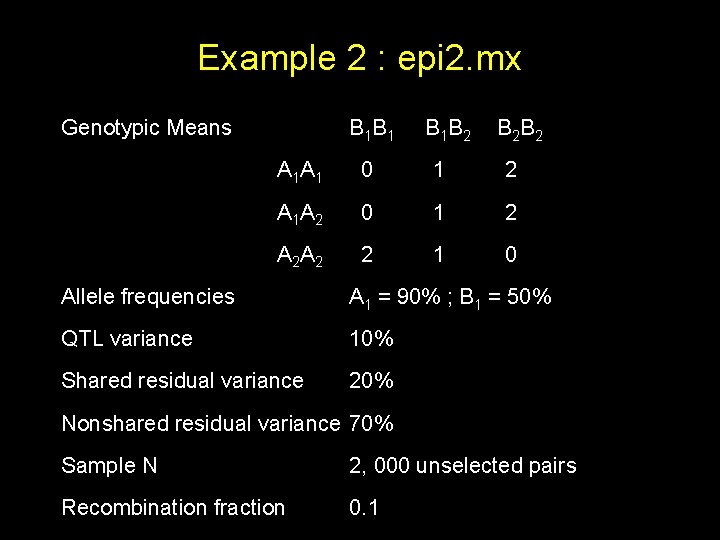
Example 2 : epi 2. mx Genotypic Means B 1 B 1 B 1 B 2 B 2 B 2 A 1 A 1 0 1 2 A 1 A 2 0 1 2 A 2 A 2 2 1 0 Allele frequencies A 1 = 90% ; B 1 = 50% QTL variance 10% Shared residual variance 20% Nonshared residual variance 70% Sample N 2, 000 unselected pairs Recombination fraction 0. 1

Exercise Using the module, are there any configurations of means, allele frequencies and recombination fraction that result in only epistatic components of variance? How does linkage between two epistatically interacting loci impact on multilocus analysis?

Poor power to detect epistasis Detection = reduction in model fit when a term is dropped Apparent variance components “soak up” variance attributable to the dropped term artificially reduces the size of the reduction

Epistasis as main effect Epistatic effects detected as additive effects “Main effect” versus “interaction effect” blurred for linkage, main effects and interaction effects are partially confounded

Probability Function Calculator http: //statgen. iop. kcl. ac. uk/bgim/ Genetic Power Calculator http: //statgen. iop. kcl. ac. uk/gpc/
 Shaun purcell
Shaun purcell Pak sham
Pak sham Smith purcell radiation
Smith purcell radiation Victoria purcell gates
Victoria purcell gates Phil purcell planned giving
Phil purcell planned giving Roland purcell a technical writer
Roland purcell a technical writer Zhang et al
Zhang et al Sikloida
Sikloida Smith purcell radiation
Smith purcell radiation Moonrise poem questions and answers
Moonrise poem questions and answers Stephen purcell
Stephen purcell Traube space
Traube space Sham peer review
Sham peer review Hala ghorani
Hala ghorani Sham dekking kat
Sham dekking kat Sham dekking kat
Sham dekking kat Sham job
Sham job Draagtijd poes
Draagtijd poes Sham al naseem
Sham al naseem Why was the cherokees last treaty a sham
Why was the cherokees last treaty a sham Dichte funktional theorie
Dichte funktional theorie Shaun harris lse
Shaun harris lse Dr michael witt
Dr michael witt Shaun topham
Shaun topham Shaun white diabetes
Shaun white diabetes Shaun moss
Shaun moss Shaun tan interesting facts
Shaun tan interesting facts Shaun loke
Shaun loke Shaun dewitt
Shaun dewitt Shaun keihn
Shaun keihn Epistasis
Epistasis Reciprocal recessive epistasis
Reciprocal recessive epistasis Duplicate dominant epistasis
Duplicate dominant epistasis Pola hereditas
Pola hereditas Mendelian
Mendelian Epistasis ejemplos
Epistasis ejemplos Epistasis
Epistasis Epistasis ejemplos
Epistasis ejemplos