Genetic Theory Pak Sham SGDP Io P London





![ACE Model for twin data 1 [0. 5/1] E C e c PT 1 ACE Model for twin data 1 [0. 5/1] E C e c PT 1](https://slidetodoc.com/presentation_image_h2/1af92a0702985367cf065eded2a97e29/image-40.jpg)
![QTL linkage model for sib-pair data 1 [0 / 0. 5 / 1] N QTL linkage model for sib-pair data 1 [0 / 0. 5 / 1] N](https://slidetodoc.com/presentation_image_h2/1af92a0702985367cf065eded2a97e29/image-41.jpg)
- Slides: 45

Genetic Theory Pak Sham SGDP, Io. P, London, UK

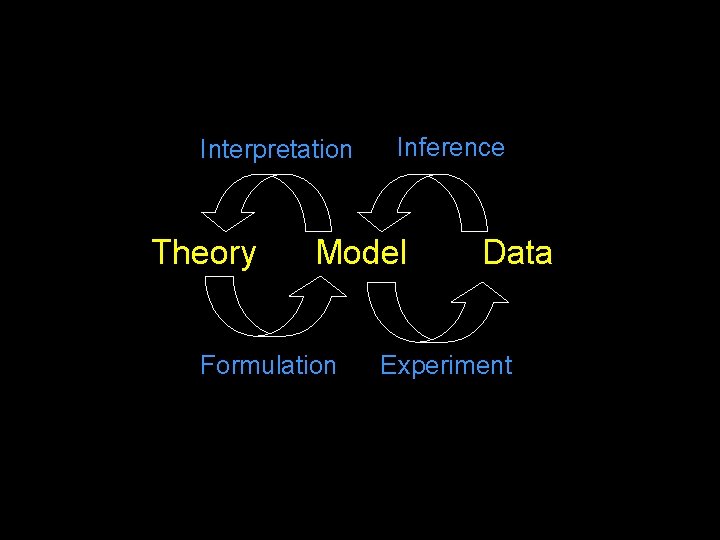
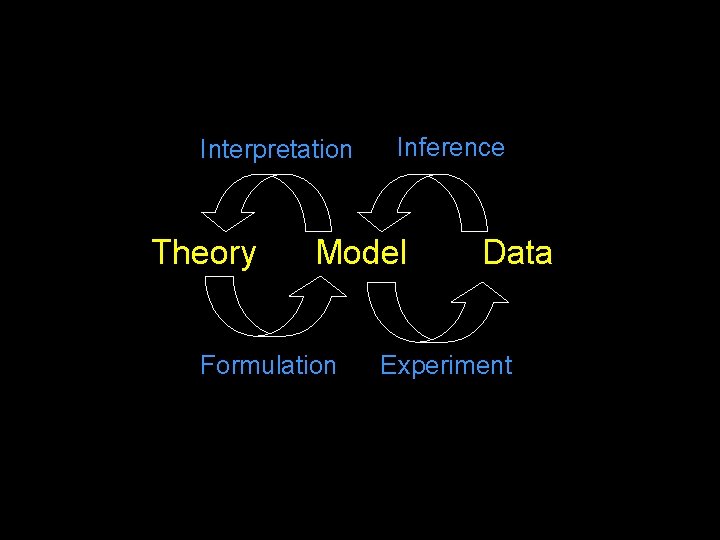
Interpretation Theory Inference Model Formulation Data Experiment

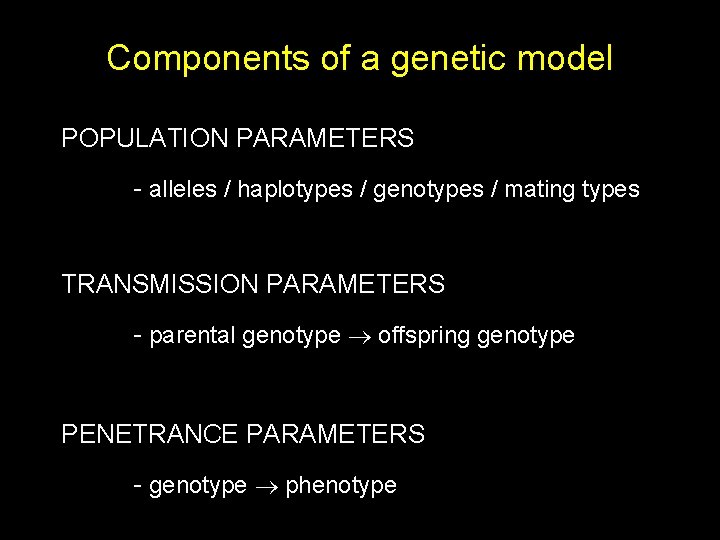
Components of a genetic model POPULATION PARAMETERS - alleles / haplotypes / genotypes / mating types TRANSMISSION PARAMETERS - parental genotype offspring genotype PENETRANCE PARAMETERS - genotype phenotype

Transmission : Mendel’s law of segregation Maternal A A ½ ½ AA ¼ AA AA ¼ ¼ ½ A Paternal A ½

Two offspring Sib 2 AA S i b 1 AA AA AA AA AA AA AA AA AA AA

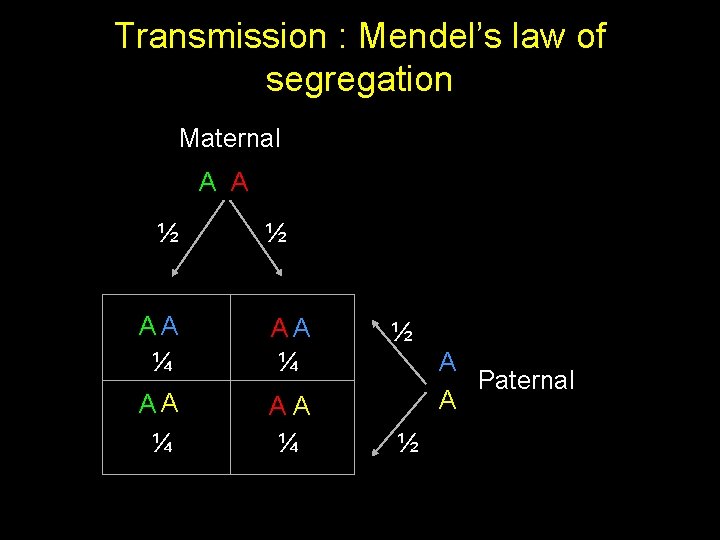
IBD sharing for two sibs AA AA AA 2 1 1 0 AA 1 2 0 1 AA 1 0 2 1 AA 0 1 1 2 Pr(IBD=0) = 4 / 16 = 0. 25 Pr(IBD=1) = 8 / 16 = 0. 50 Pr(IBD=2) = 4 / 16 = 0. 25 Expected IBD sharing = (2*0. 25) + (1*0. 5) + (0*0. 25) =1

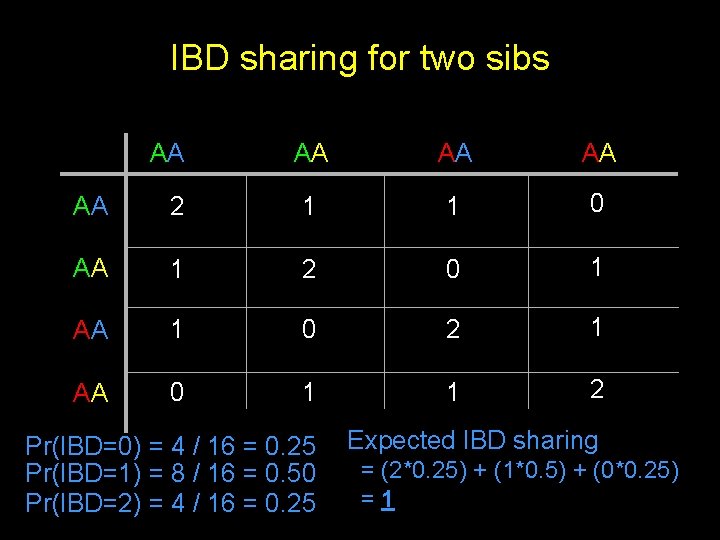
IBS IBD A 1 A 2 A 1 A 3 IBS = 1 IBD = 0 A 1 A 2 A 1 A 3

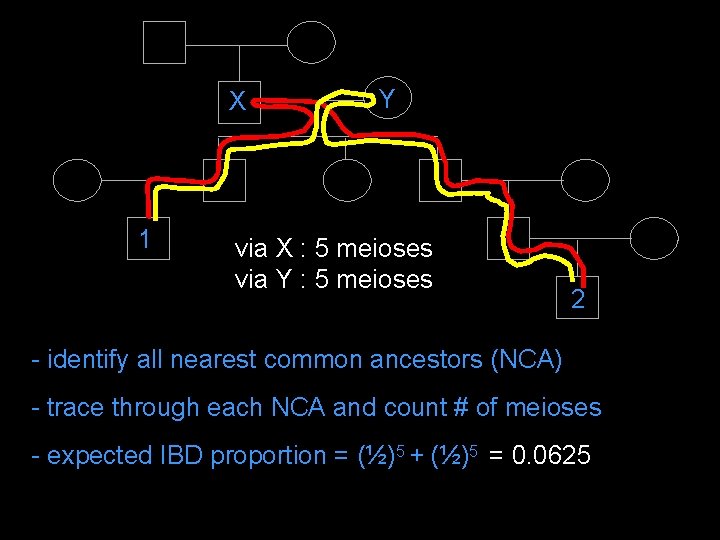
X 1 Y via X : 5 meioses via Y : 5 meioses 2 - identify all nearest common ancestors (NCA) - trace through each NCA and count # of meioses - expected IBD proportion = (½)5 + (½)5 = 0. 0625

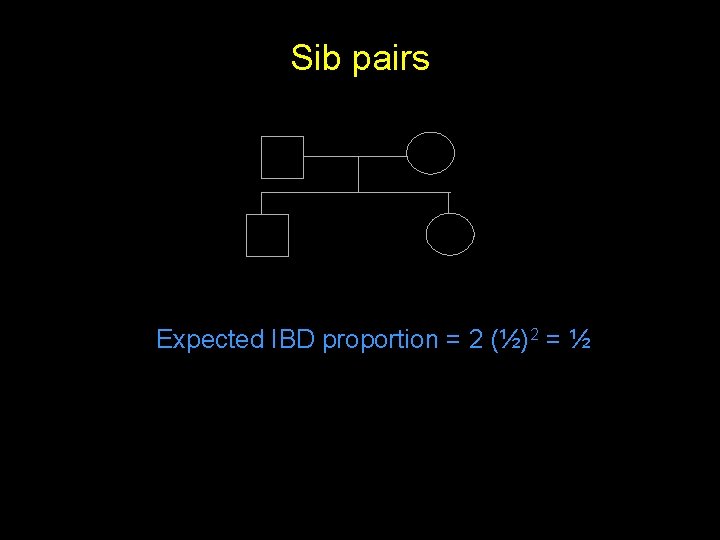
Sib pairs Expected IBD proportion = 2 (½)2 = ½

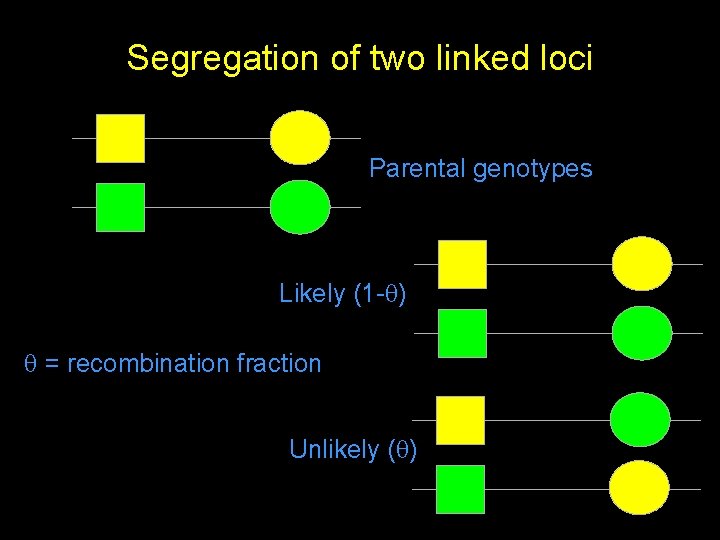
Segregation of two linked loci Parental genotypes Likely (1 - ) = recombination fraction Unlikely ( )

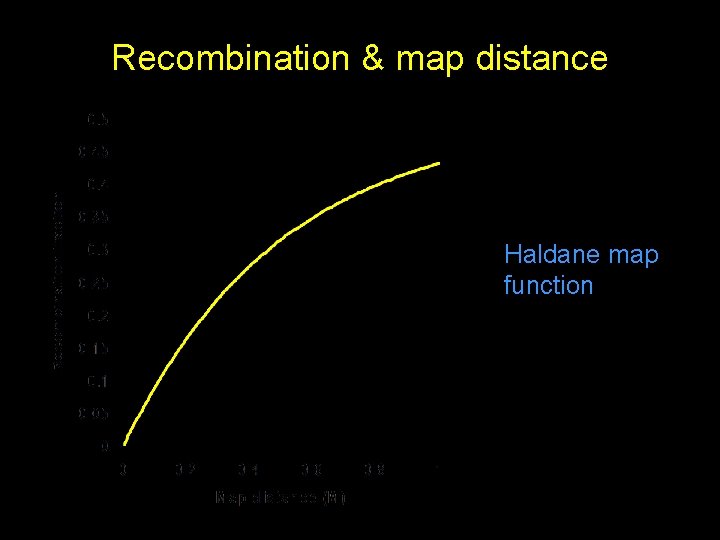
Recombination & map distance Haldane map function

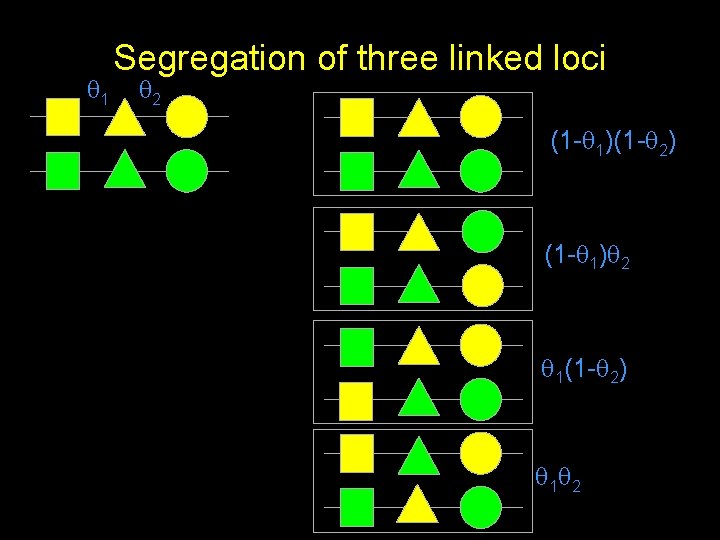
1 Segregation of three linked loci 2 (1 - 1)(1 - 2) (1 - 1) 2 1(1 - 2) 1 2

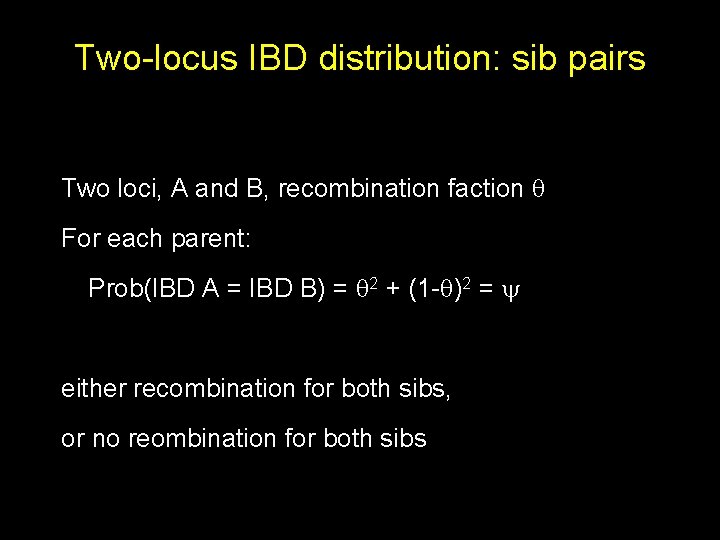
Two-locus IBD distribution: sib pairs Two loci, A and B, recombination faction For each parent: Prob(IBD A = IBD B) = 2 + (1 - )2 = either recombination for both sibs, or no reombination for both sibs

Conditional distribution of at maker given at QTL at M 0 1/2 1

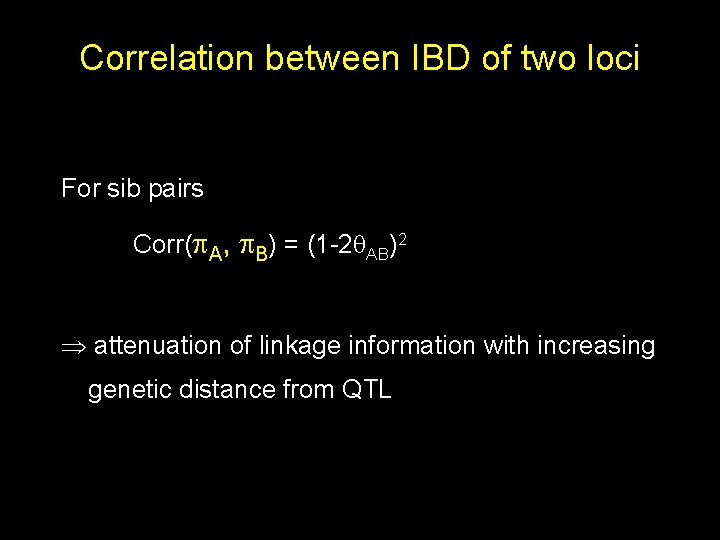
Correlation between IBD of two loci For sib pairs Corr( A, B) = (1 -2 AB)2 attenuation of linkage information with increasing genetic distance from QTL

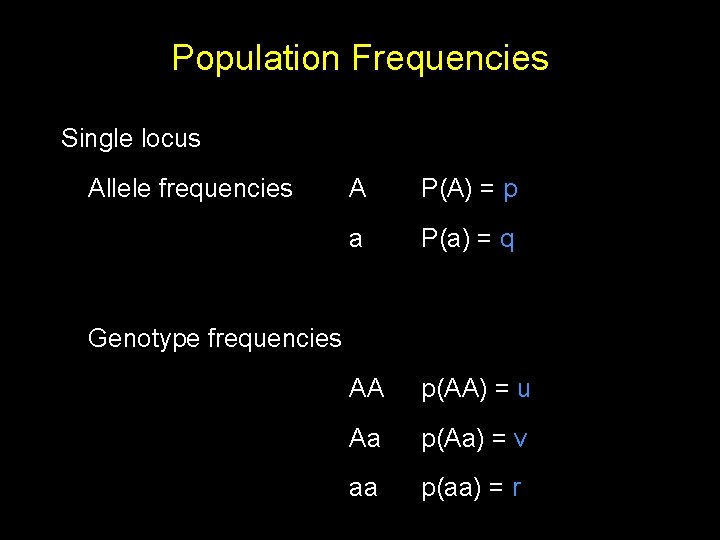
Population Frequencies Single locus Allele frequencies A P(A) = p a P(a) = q AA p(AA) = u Aa p(Aa) = v aa p(aa) = r Genotype frequencies

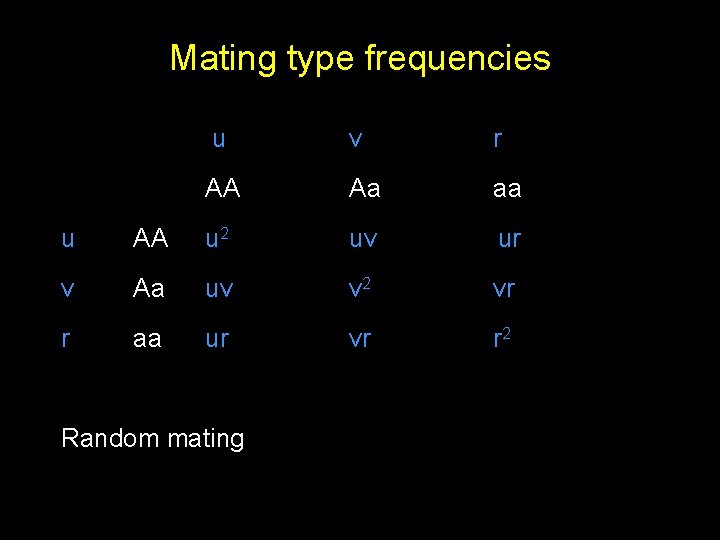
Mating type frequencies u v r AA Aa aa u AA u 2 uv ur v Aa uv v 2 vr r aa ur vr r 2 Random mating

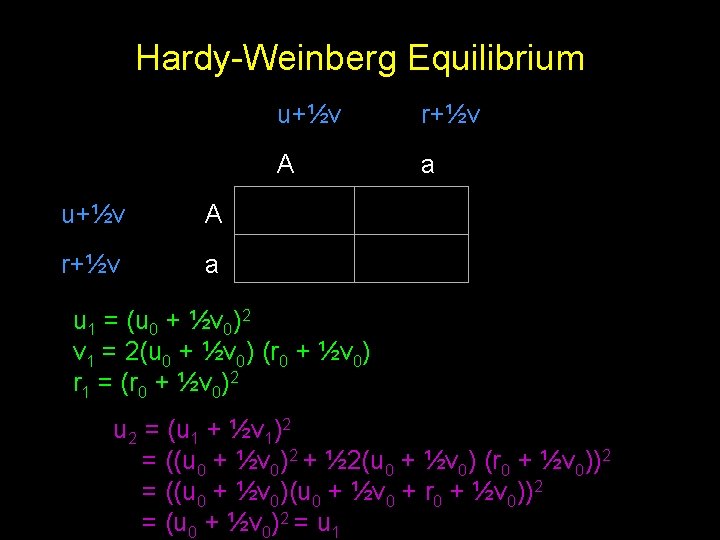
Hardy-Weinberg Equilibrium u+½v A r+½v a u+½v r+½v A a u 1 = (u 0 + ½v 0)2 v 1 = 2(u 0 + ½v 0) (r 0 + ½v 0) r 1 = (r 0 + ½v 0)2 u 2 = (u 1 + ½v 1)2 = ((u 0 + ½v 0)2 + ½ 2(u 0 + ½v 0) (r 0 + ½v 0))2 = ((u 0 + ½v 0)(u 0 + ½v 0 + r 0 + ½v 0))2 = (u 0 + ½v 0)2 = u 1

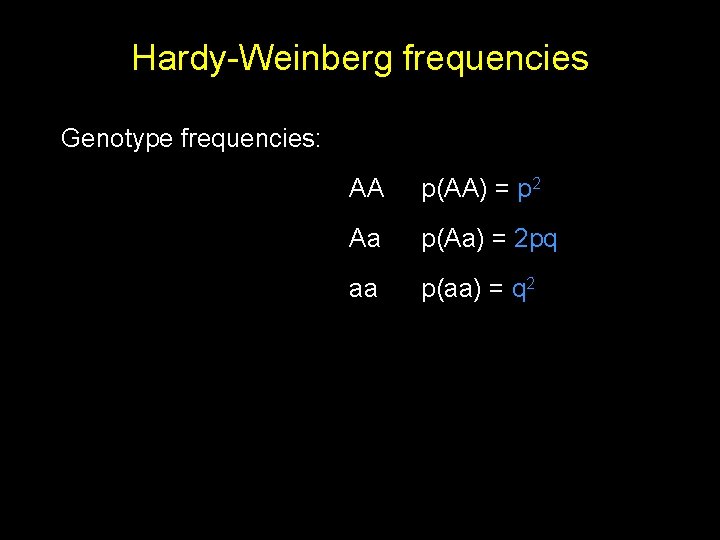
Hardy-Weinberg frequencies Genotype frequencies: AA p(AA) = p 2 Aa p(Aa) = 2 pq aa p(aa) = q 2

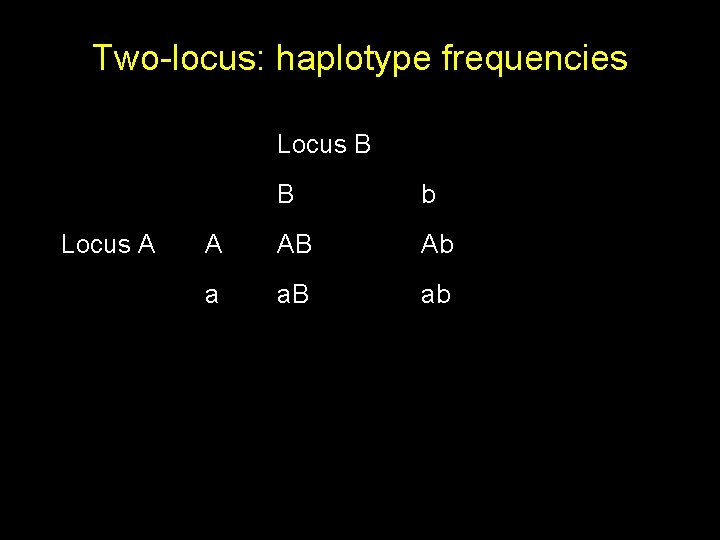
Two-locus: haplotype frequencies Locus B Locus A B b A AB Ab a a. B ab

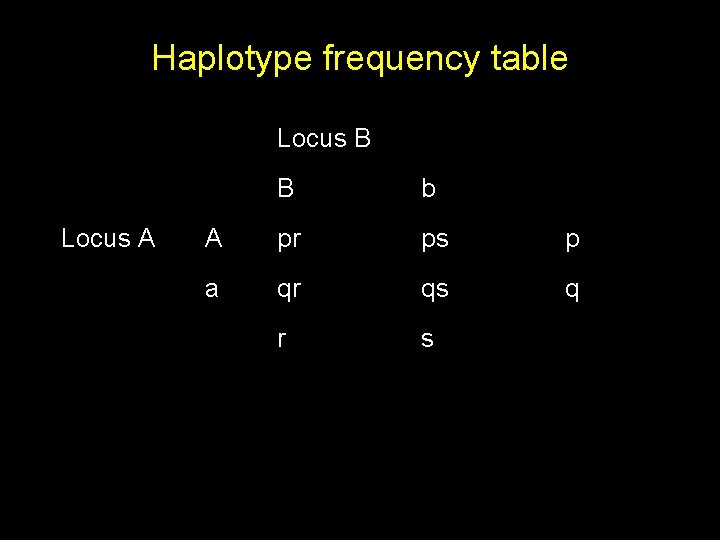
Haplotype frequency table Locus B Locus A B b A pr ps p a qr qs q r s

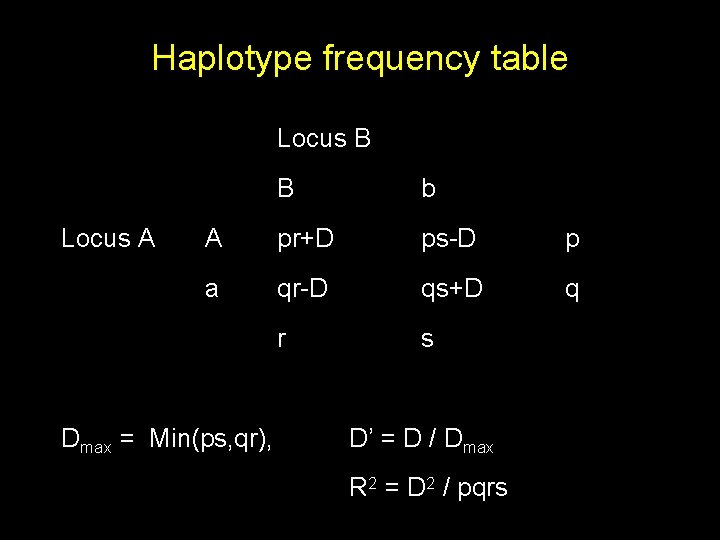
Haplotype frequency table Locus B Locus A B b A pr+D ps-D p a qr-D qs+D q r s Dmax = Min(ps, qr), D’ = D / Dmax R 2 = D 2 / pqrs

Causes of allelic association Tight Linkage Founder effect: D (1 - )G Genetic Drift: R 2 (NE )-1 Population admixture Selection

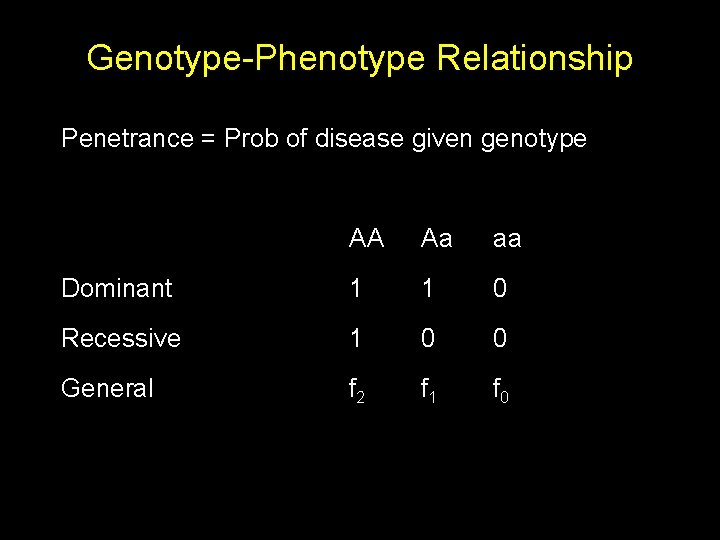
Genotype-Phenotype Relationship Penetrance = Prob of disease given genotype AA Aa aa Dominant 1 1 0 Recessive 1 0 0 General f 2 f 1 f 0

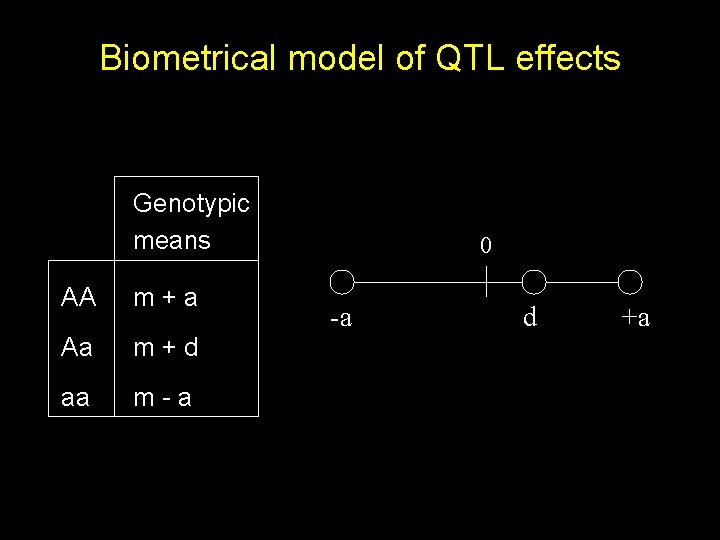
Biometrical model of QTL effects Genotypic means AA m+a Aa m+d aa m-a 0 -a d +a

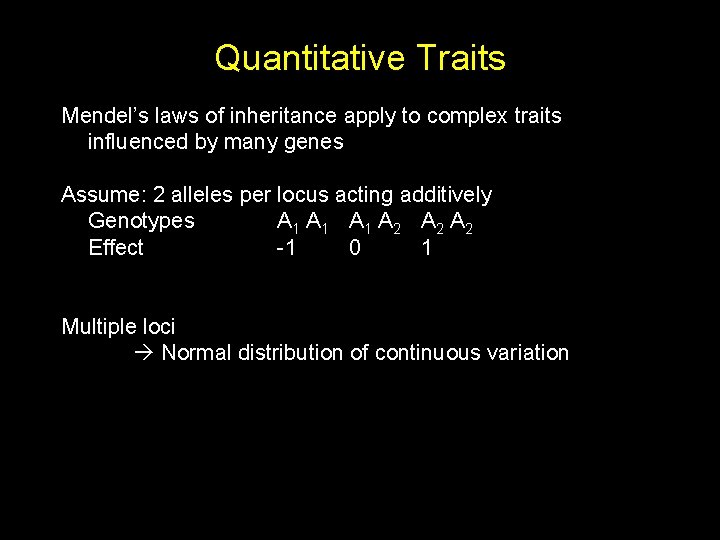
Quantitative Traits Mendel’s laws of inheritance apply to complex traits influenced by many genes Assume: 2 alleles per locus acting additively Genotypes A 1 A 1 A 2 A 2 Effect -1 0 1 Multiple loci Normal distribution of continuous variation

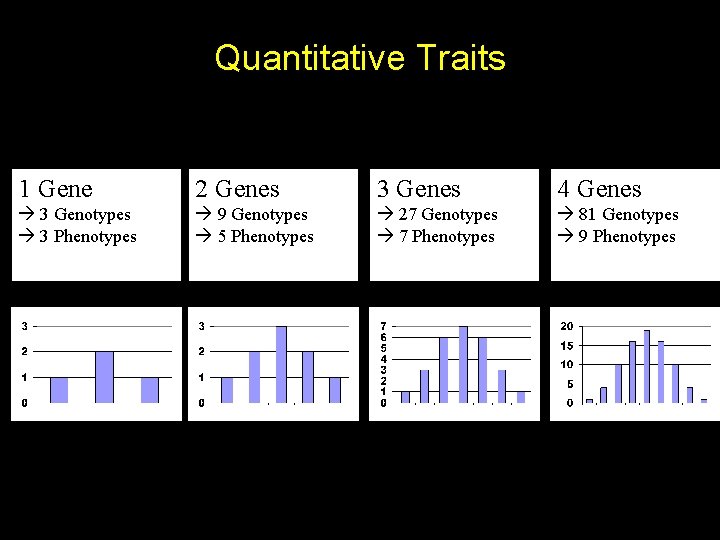
Quantitative Traits 1 Gene 2 Genes 3 Genes 4 Genes 3 Genotypes 3 Phenotypes 9 Genotypes 5 Phenotypes 27 Genotypes 7 Phenotypes 81 Genotypes 9 Phenotypes

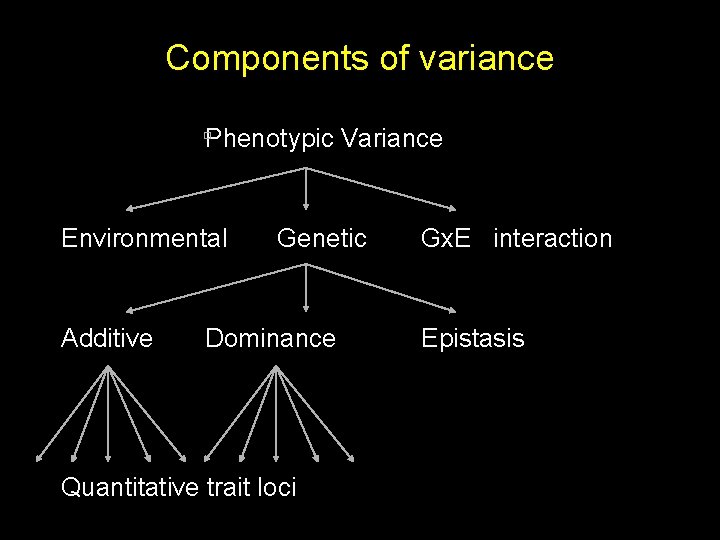
Components of variance Phenotypic Variance Environmental Genetic Gx. E interaction

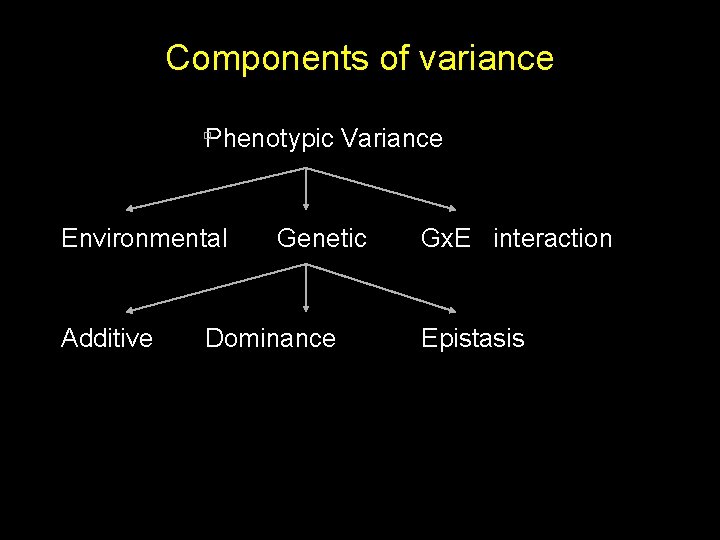
Components of variance Phenotypic Variance Environmental Additive Genetic Dominance Gx. E interaction Epistasis

Components of variance Phenotypic Variance Environmental Additive Genetic Dominance Quantitative trait loci Gx. E interaction Epistasis

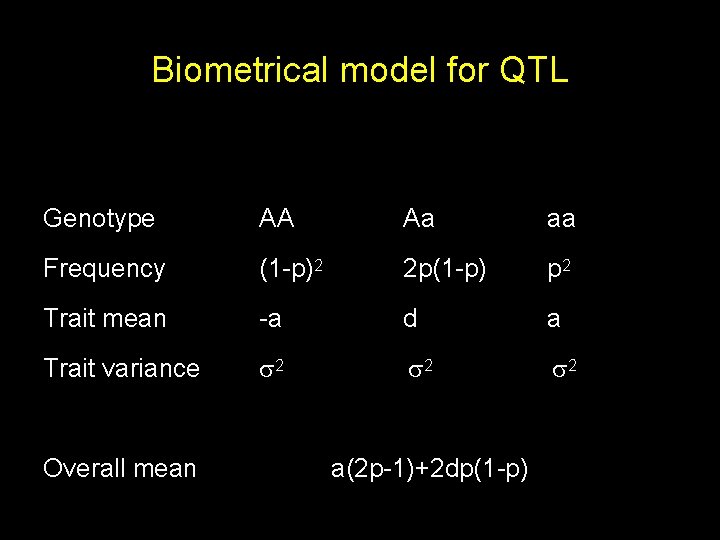
Biometrical model for QTL Genotype AA Aa aa Frequency (1 -p)2 2 p(1 -p) p 2 Trait mean -a d a Trait variance 2 2 2 Overall mean a(2 p-1)+2 dp(1 -p)

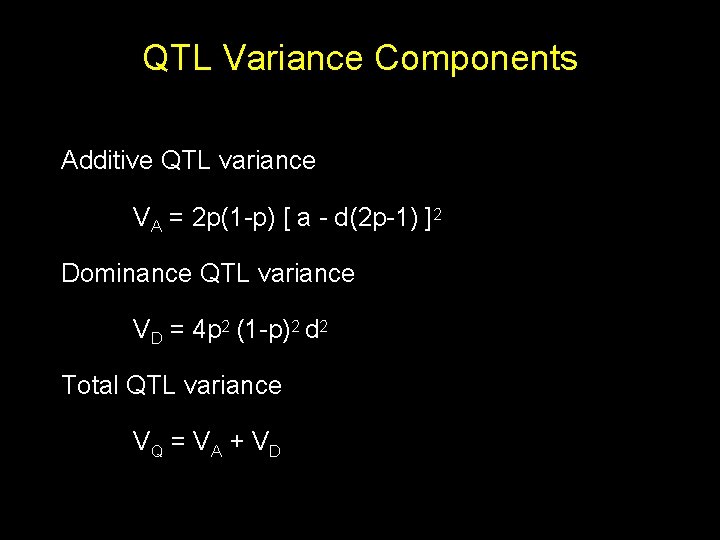
QTL Variance Components Additive QTL variance VA = 2 p(1 -p) [ a - d(2 p-1) ]2 Dominance QTL variance VD = 4 p 2 (1 -p)2 d 2 Total QTL variance VQ = V A + V D

Covariance between relatives Partition of variance Partition of covariance Overall covariance = sum of covariances of all components Covariance of component between relatives = correlation of component variance due to component

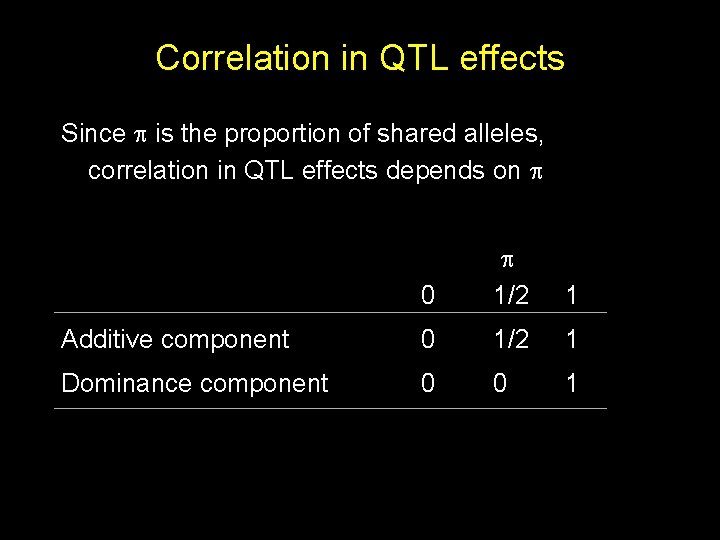
Correlation in QTL effects Since is the proportion of shared alleles, correlation in QTL effects depends on 0 1/2 1 Additive component 0 1/2 1 Dominance component 0 0 1

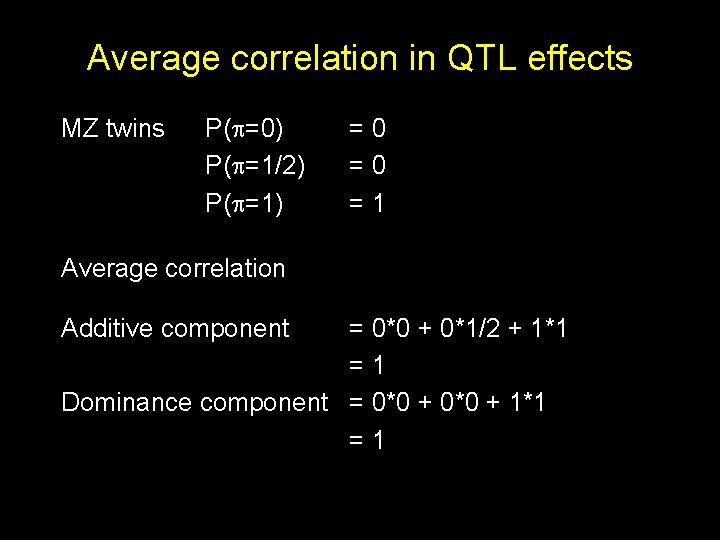
Average correlation in QTL effects MZ twins P( =0) P( =1/2) P( =1) =0 =0 =1 Average correlation Additive component = 0*0 + 0*1/2 + 1*1 =1 Dominance component = 0*0 + 1*1 =1

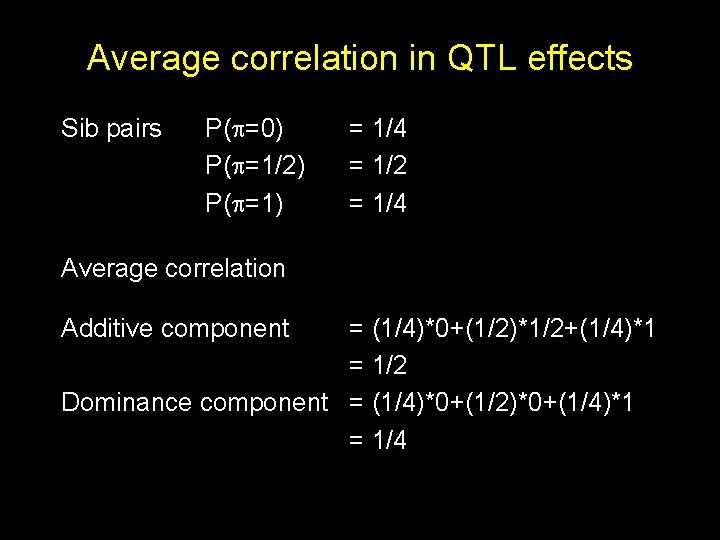
Average correlation in QTL effects Sib pairs P( =0) P( =1/2) P( =1) = 1/4 = 1/2 = 1/4 Average correlation Additive component = (1/4)*0+(1/2)*1/2+(1/4)*1 = 1/2 Dominance component = (1/4)*0+(1/2)*0+(1/4)*1 = 1/4

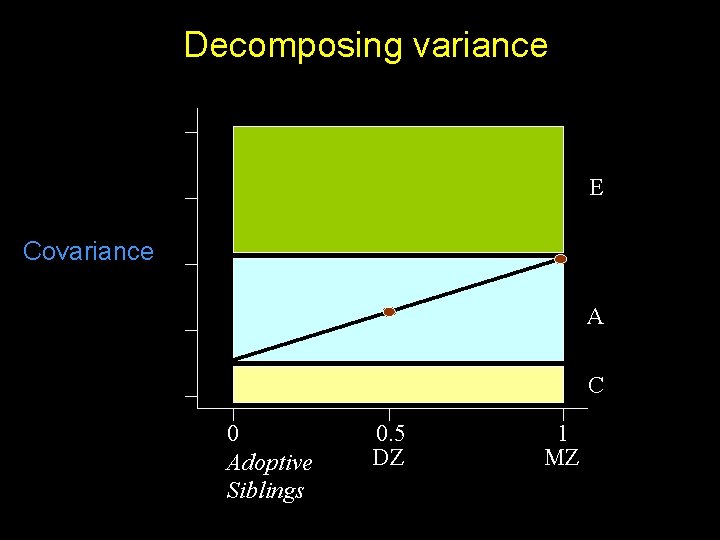
Decomposing variance E Covariance A C 0 Adoptive Siblings 0. 5 DZ 1 MZ

Path analysis allows us to diagrammatically represent linear models for the relationships between variables easy to derive expectations for the variances and covariances of variables in terms of the parameters of the proposed linear model permits translation into matrix formulation (Mx)

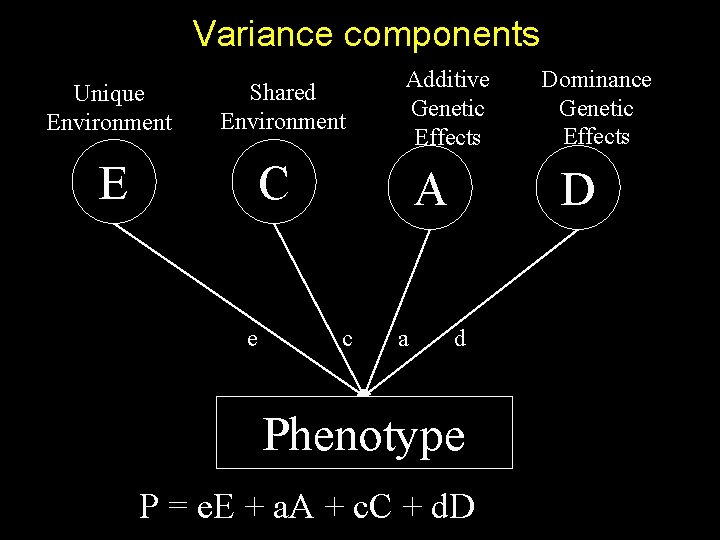
Variance components Unique Environment Shared Environment E Additive Genetic Effects C e A c a Dominance Genetic Effects D d Phenotype P = e. E + a. A + c. C + d. D
![ACE Model for twin data 1 0 51 E C e c PT 1 ACE Model for twin data 1 [0. 5/1] E C e c PT 1](https://slidetodoc.com/presentation_image_h2/1af92a0702985367cf065eded2a97e29/image-40.jpg)
ACE Model for twin data 1 [0. 5/1] E C e c PT 1 A a A C a c PT 2 E e
![QTL linkage model for sibpair data 1 0 0 5 1 N QTL linkage model for sib-pair data 1 [0 / 0. 5 / 1] N](https://slidetodoc.com/presentation_image_h2/1af92a0702985367cf065eded2a97e29/image-41.jpg)
QTL linkage model for sib-pair data 1 [0 / 0. 5 / 1] N S n s PT 1 Q q Q S q s PT 2 N n

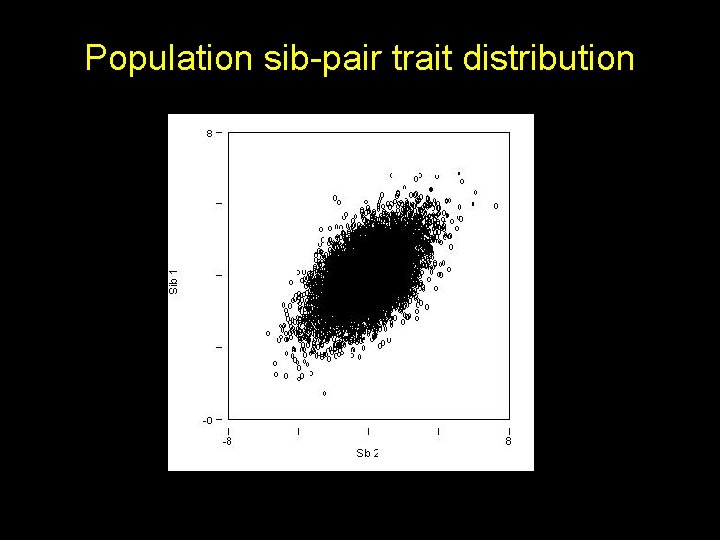
Population sib-pair trait distribution

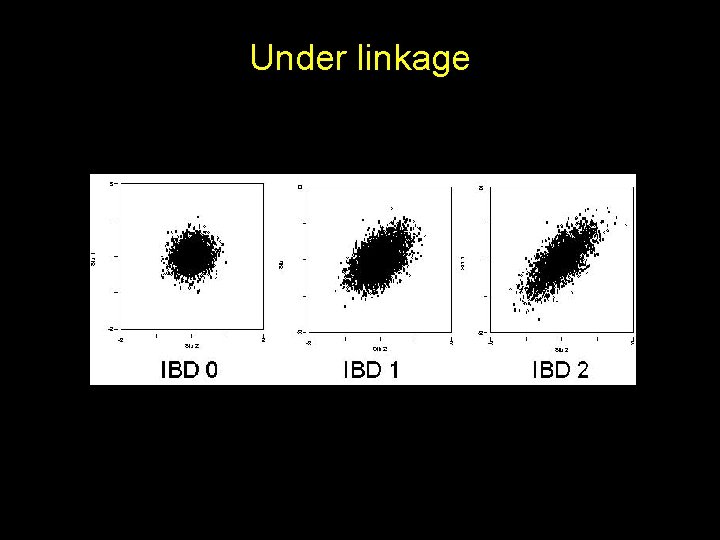
Under linkage

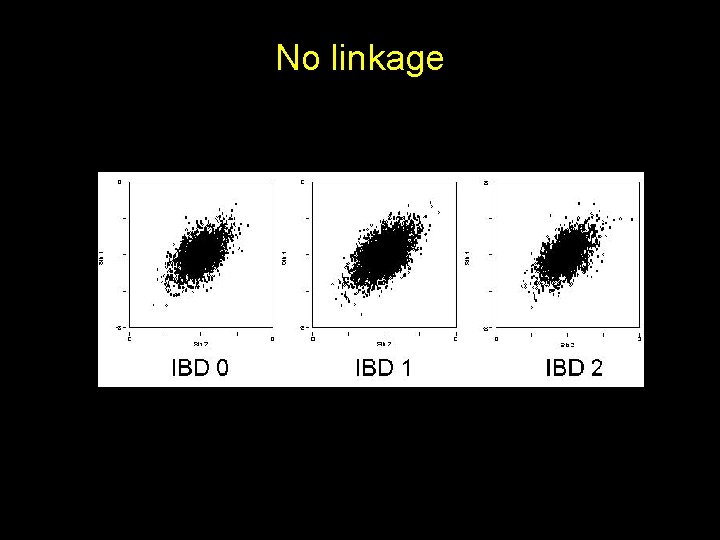
No linkage

Interpretation Theory Inference Model Formulation Data Experiment
 Pak sham
Pak sham How is genetic drift different from gene flow
How is genetic drift different from gene flow Genetic programming vs genetic algorithm
Genetic programming vs genetic algorithm Genetic programming vs genetic algorithm
Genetic programming vs genetic algorithm Gene flow vs genetic drift
Gene flow vs genetic drift Genetic drift vs genetic flow
Genetic drift vs genetic flow Stopt krolsheid na dekking
Stopt krolsheid na dekking Kat ce
Kat ce About how many cherokees died of smallpox in 1745
About how many cherokees died of smallpox in 1745 Sham peer review
Sham peer review Bronchial vs vesicular breathing
Bronchial vs vesicular breathing Poezenpil na bevalling
Poezenpil na bevalling Sham al naseem
Sham al naseem Dft dichte funktional theorie
Dft dichte funktional theorie Sham country map
Sham country map Sham job
Sham job Biological trait theory criminology
Biological trait theory criminology Dairy products packaging machine
Dairy products packaging machine Pak solhin
Pak solhin Tetra pak taiwan
Tetra pak taiwan Pak ramlan mempunyai rumah dan kebun njop
Pak ramlan mempunyai rumah dan kebun njop Perbandingan trigonometri segitiga siku siku
Perbandingan trigonometri segitiga siku siku Dupak pengawas sekolah
Dupak pengawas sekolah Tetrapak orbis
Tetrapak orbis Ciri-ciri pak 21
Ciri-ciri pak 21 Pak amin membeli beras dari cianjur
Pak amin membeli beras dari cianjur Predatory pricing
Predatory pricing Kekuatan dan jenis kacamata pak anto adalah
Kekuatan dan jenis kacamata pak anto adalah Tetra pak france organigramme
Tetra pak france organigramme Tehno eko pak
Tehno eko pak Dari hasil tes ternyata urin pak yudha mengandung glukosa
Dari hasil tes ternyata urin pak yudha mengandung glukosa Desti mendapat penghasilan sebesar rp900.000
Desti mendapat penghasilan sebesar rp900.000 Keuntungan yang diperoleh pak karta semakin bertambah
Keuntungan yang diperoleh pak karta semakin bertambah Life pak +
Life pak + Sebuah perusahaan percetakan memberikan rabat 30
Sebuah perusahaan percetakan memberikan rabat 30 Zweetpak
Zweetpak Contoh unsur penunjang
Contoh unsur penunjang Warna merah cerah pada darah manusia disebabkan oleh
Warna merah cerah pada darah manusia disebabkan oleh Renungan ulangan 6 4-9
Renungan ulangan 6 4-9 Pertanyaan tentang pengolahan skor
Pertanyaan tentang pengolahan skor Pak naw
Pak naw Pak bowo pengrajin wayang golek
Pak bowo pengrajin wayang golek Robert pak
Robert pak Access quarterly packages
Access quarterly packages Anghkar
Anghkar Umur pak agus 3 kali umur iwan
Umur pak agus 3 kali umur iwan