QTL and QTL allele validation in cherry Amy

















- Slides: 43

QTL and QTL allele validation in cherry Amy Iezzoni Cameron Peace, Audrey Sebolt, Nnadozie Oraguzie, Umesh Rosyara, Travis Stegmeir 25 July 2013 ASHS Palm Desert, CA

Outline of Presentation v What is QTL Validation? What is QTL allele Validation? v QTL Validation: FW_G 2 for Fruit Size v QTL Allele Validation for FW_G 2 v Other Jewels for Cherry

What is QTL Validation? What is QTL Allele Validation?

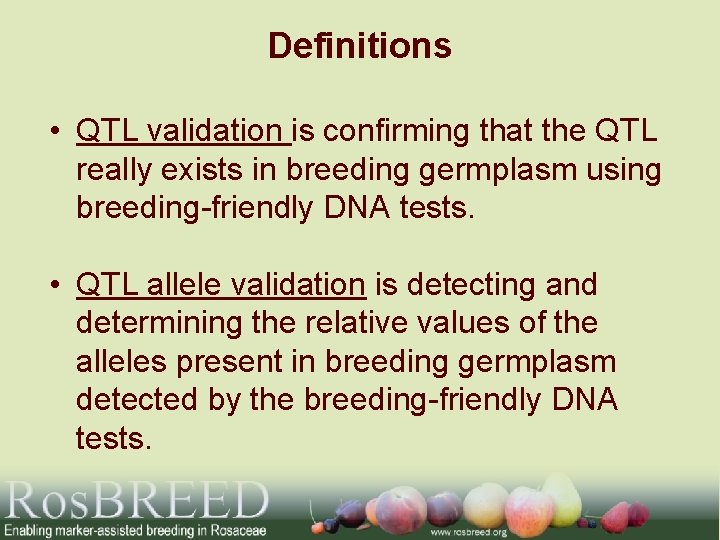
Definitions • QTL validation is confirming that the QTL really exists in breeding germplasm using breeding-friendly DNA tests. • QTL allele validation is detecting and determining the relative values of the alleles present in breeding germplasm detected by the breeding-friendly DNA tests.

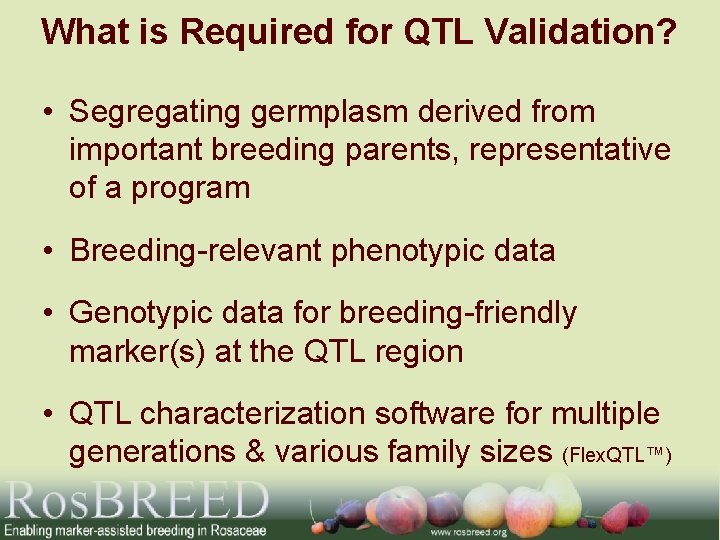
What is Required for QTL Validation? • Segregating germplasm derived from important breeding parents, representative of a program • Breeding-relevant phenotypic data • Genotypic data for breeding-friendly marker(s) at the QTL region • QTL characterization software for multiple generations & various family sizes (Flex. QTL™)

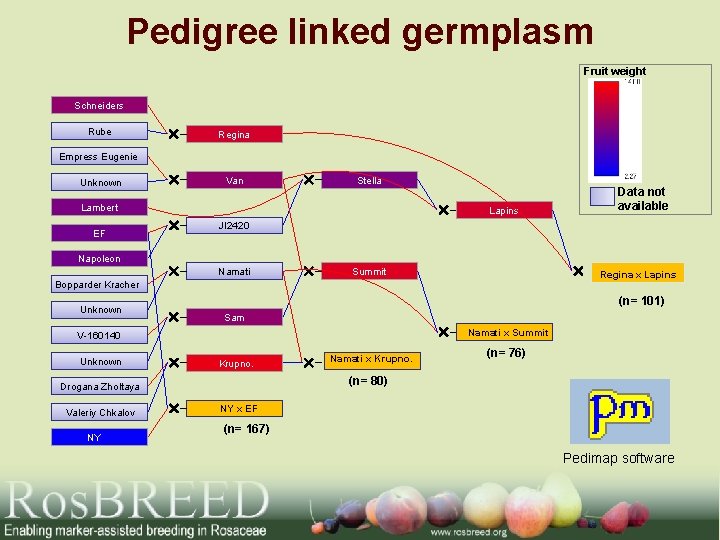
Pedigree linked germplasm Fruit weight Schneiders Rube Regina Empress Eugenie Unknown Van Stella Lambert EF Lapins Data not available JI 2420 Napoleon Namati Summit Regina x Lapins Bopparder Kracher Unknown (n= 101) Sam Namati x Summit V-160140 Unknown Krupno. NY (n= 76) (n= 80) Drogana Zholtaya Valeriy Chkalov Namati x Krupno. NY x EF (n= 167) Pedimap software

Standardized Phenotyping • Reference Germplasm Sets • Standardized phenotyping at multiple locations, esp. for fruit quality (www. rosbreed. org) • Evaluations done for 2 – 3 years • Available at Genome Database for Rosaceae (www. rosaceae. org/breeders_toolbox) Ros. BREED

Genome Wide SNP arrays • Genome-scanning SNP arrays developed and utilized for apple (9 K), peach (9 K) and cherry (6 K) by international Ros. BREED-led efforts

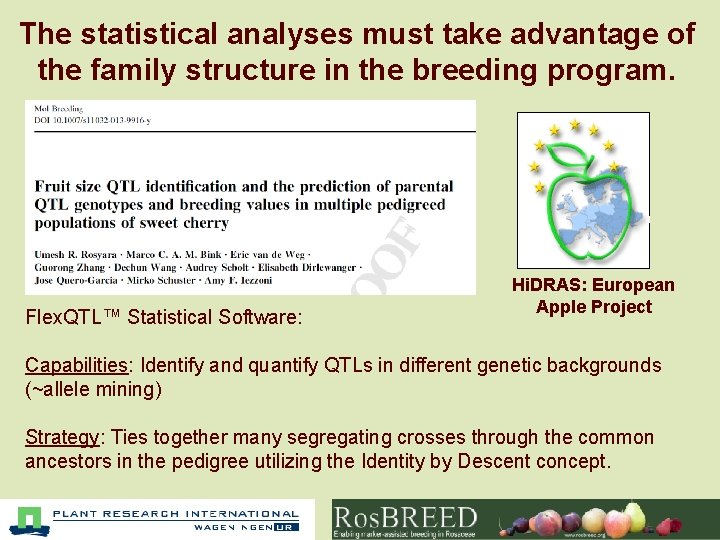
The statistical analyses must take advantage of the family structure in the breeding program. Flex. QTL™ Statistical Software: Hi. DRAS: European Apple Project Capabilities: Identify and quantify QTLs in different genetic backgrounds (~allele mining) Strategy: Ties together many segregating crosses through the common ancestors in the pedigree utilizing the Identity by Descent concept.

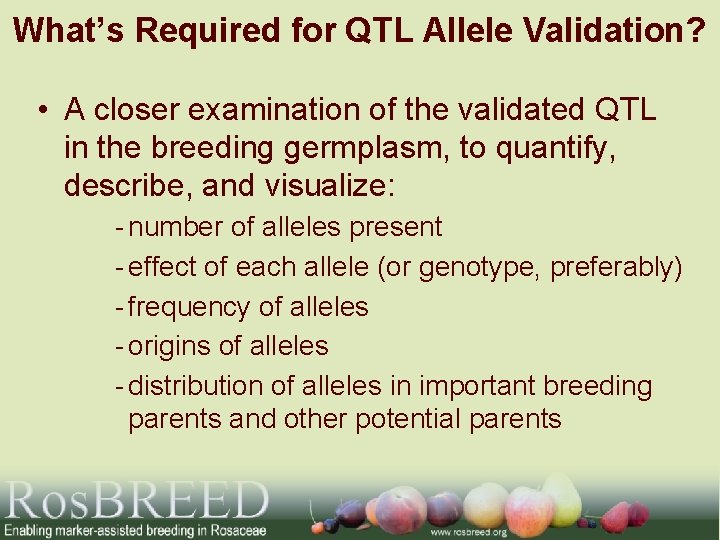
What’s Required for QTL Allele Validation? • A closer examination of the validated QTL in the breeding germplasm, to quantify, describe, and visualize: - number of alleles present - effect of each allele (or genotype, preferably) - frequency of alleles - origins of alleles - distribution of alleles in important breeding parents and other potential parents

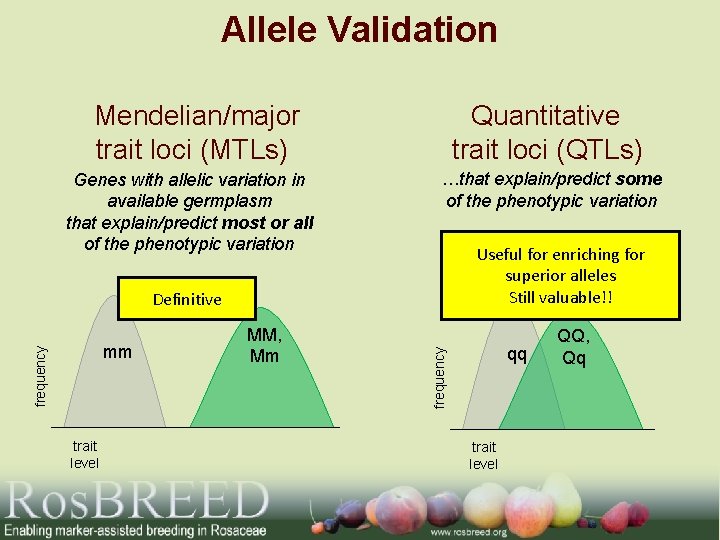
Allele Validation Mendelian/major Quantitative trait loci (MTLs) trait loci (QTLs) Genes with allelic variation in available germplasm that explain/predict most or all of the phenotypic variation …that explain/predict some of the phenotypic variation Useful for enriching for superior alleles Still valuable!! frequency mm trait level MM, Mm qq frequency Definitive trait level QQ, Qq

QTL Validation - confirming the FW_G 2 QTL really exists in cherry breeding germplasm

Example of a Valuable QTL Discovered: FW_G 2 for fruit size (Zhang et al. 2010) • Trait has value to stakeholders • QTL explains a significant amount of trait variation • Achieving the desired phenotype with breeding is very difficult

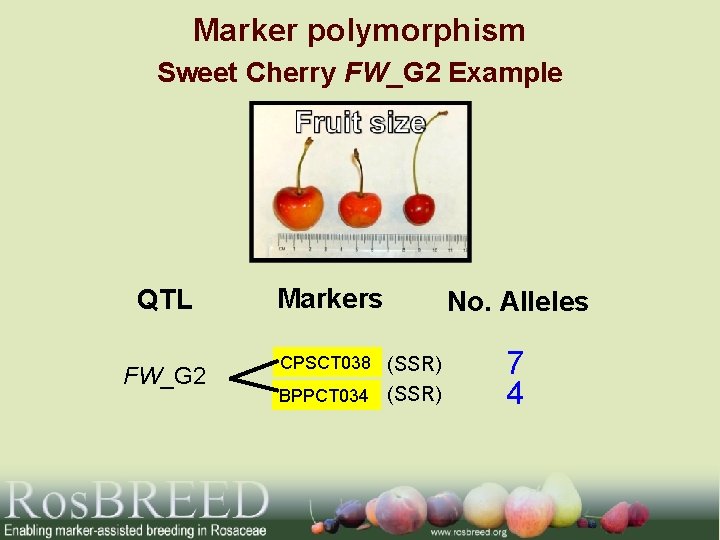
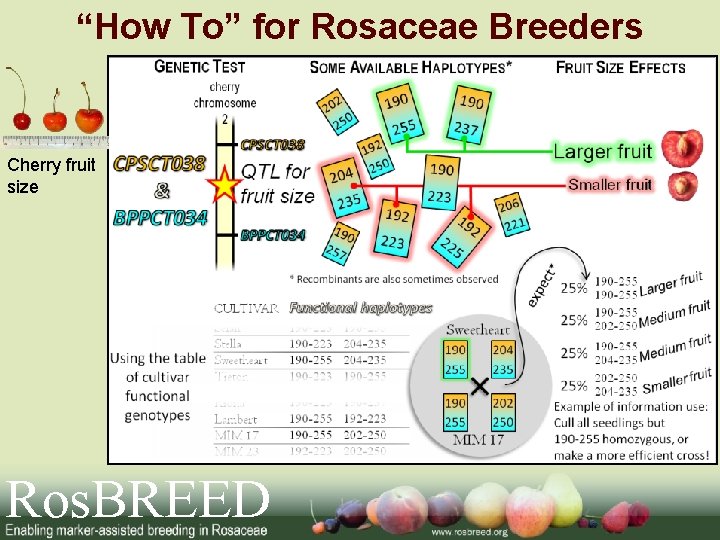
Breeding-Friendly DNA Markers Used 2 SSRs • CPSCT 038 • BPPCT 034 These 2 SSRs defined 3 functional alleles for FW_G 2 in the bi-parental cross where the QTL was discovered

Validation: Sweet Cherry Example Utility assessment on fruiting seedlings 22 populations (219 seedlings), 0 cultivars

Simple Validation and Functional Genotype Effects for the Sweet Cherry Success Story Phenotypic data collection www. rosbreed. org/resources/fruit-evaluation

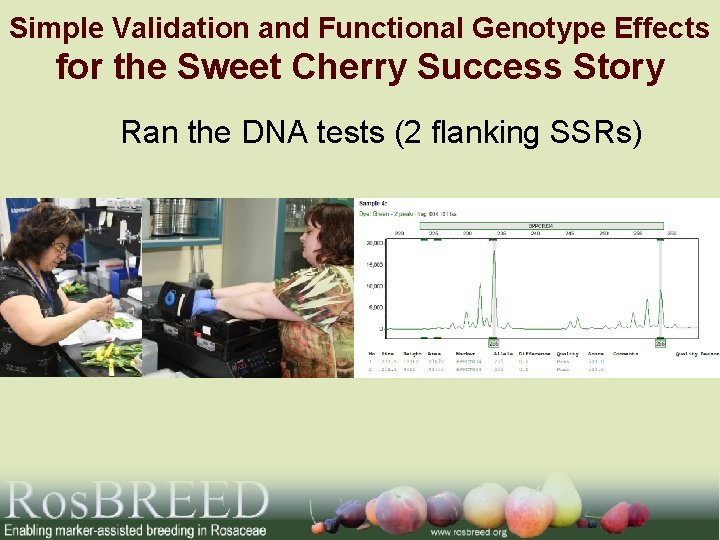
Simple Validation and Functional Genotype Effects for the Sweet Cherry Success Story Ran the DNA tests (2 flanking SSRs)

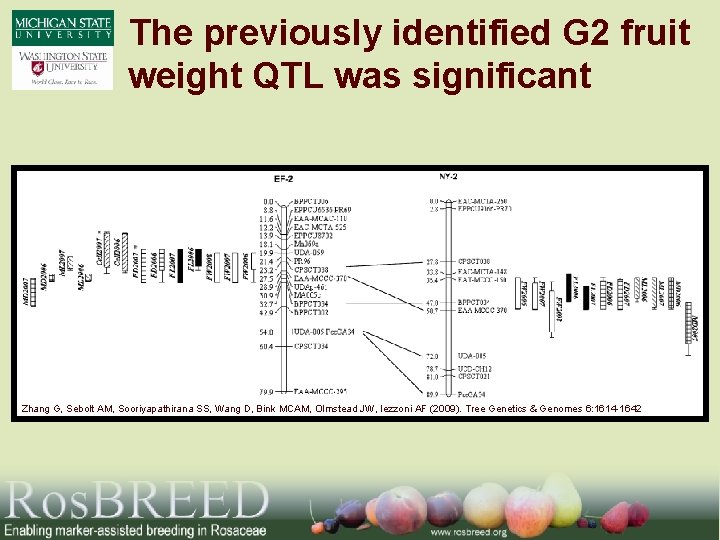
The previously identified G 2 fruit weight QTL was significant Zhang G, Sebolt AM, Sooriyapathirana SS, Wang D, Bink MCAM, Olmstead JW, Iezzoni AF (2009). Tree Genetics & Genomes 6: 1614 -1642

Marker polymorphism Sweet Cherry FW_G 2 Example QTL FW_G 2 Markers No. Alleles CPSCT 038 (SSR) BPPCT 034 (SSR) 7 4

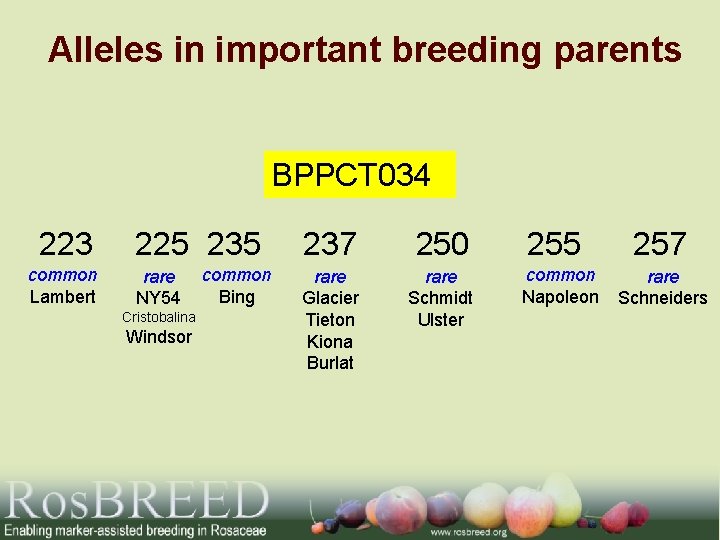
Alleles in important breeding parents BPPCT 034 223 common Lambert 225 235 rare NY 54 Cristobalina Windsor common Bing 237 250 rare Glacier Tieton Kiona Burlat rare Schmidt Ulster 255 common Napoleon 257 rare Schneiders

QTL Validation - Summary • The G 2 QTL for fruit size was identified in sweet cherry breeding germplasm • The DNA test using the flanking SSR markers was still associated with the trait • Origin and distribution of alleles in important breeding parents were determined

QTL Allele Validation - detecting and valuing FW_G 2 alleles in cherry breeding germplasm

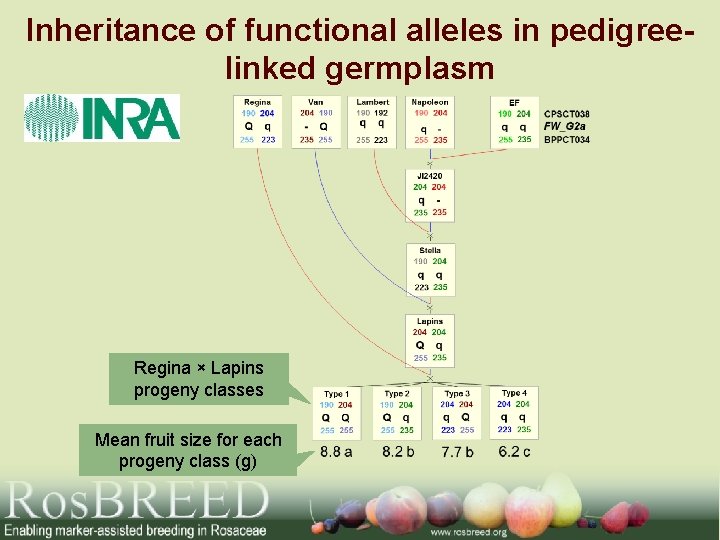
Inheritance of functional alleles in pedigreelinked germplasm Regina × Lapins progeny classes Mean fruit size for each progeny class (g)

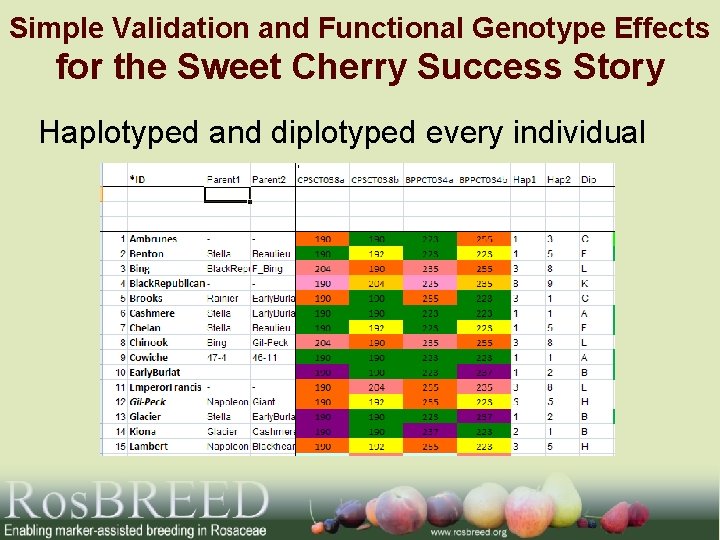
Simple Validation and Functional Genotype Effects for the Sweet Cherry Success Story Haplotyped and diplotyped every individual

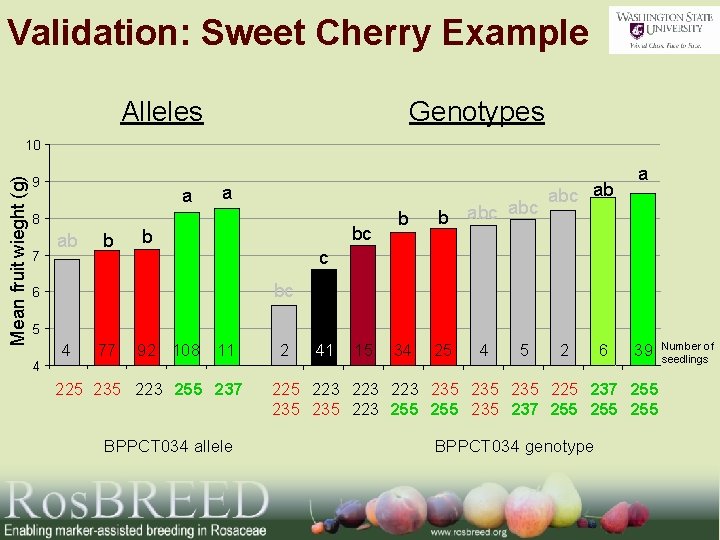
Validation: Sweet Cherry Example Alleles Genotypes Mean fruit wieght (g) 10 9 a 8 7 ab b a bc b b abc 34 25 abc ab a c bc 6 5 4 4 77 92 108 11 225 235 223 255 237 BPPCT 034 allele 2 41 15 4 5 2 6 39 225 223 223 235 235 225 237 255 235 223 255 235 237 255 255 BPPCT 034 genotype Number of seedlings

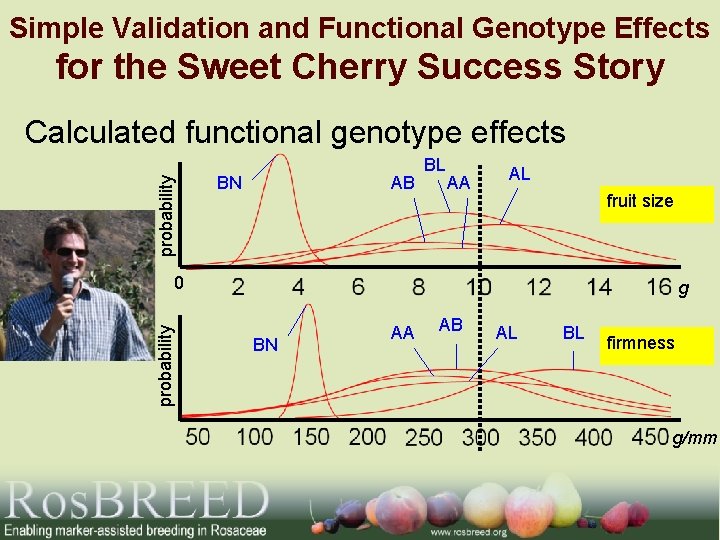
Simple Validation and Functional Genotype Effects for the Sweet Cherry Success Story probability Calculated functional genotype effects BN AB BL AA AL fruit size probability 0 g BN AA AB AL BL firmness g/mm

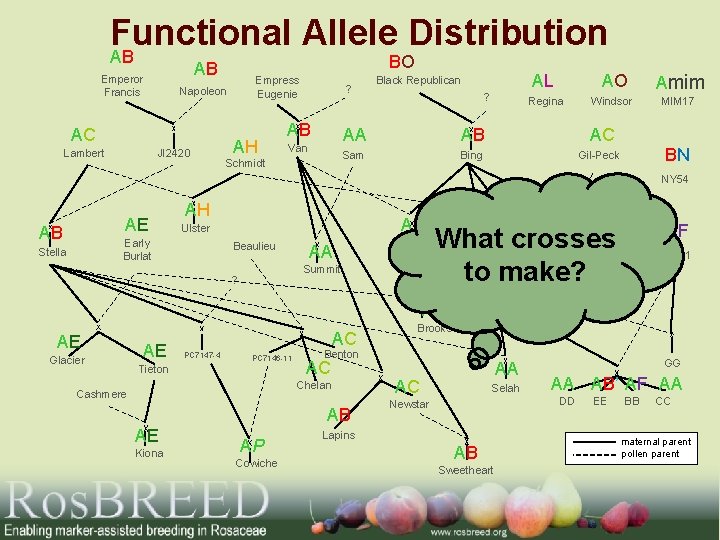
Functional Allele Distribution AB Empress Eugenie Napoleon AC Lambert BO AB Emperor Francis JI 2420 AH ? AB Van Schmidt Black Republican ? AL AO Amim Regina Windsor MIM 17 AA AB AC Sam Bing Gil-Peck BN NY 54 AE AB AH Early Burlat Stella AB Ulster Beaulieu AA Rainier Summit ? AE Glacier AE PC 7147 -4 PC 7146 -11 Tieton Chelan Cashmere AB AE Kiona AP Cowiche Vic Chinook AF PMR-1 P 8 -79 Brooks Benton AC AB What crosses to make? AA AC AA AA AC Selah GG AA AB’ AF AA DD Newstar Lapins AB Sweetheart EE BB CC maternal parent pollen parent

“How To” for Rosaceae Breeders Cherry fruit size Ros. BREED

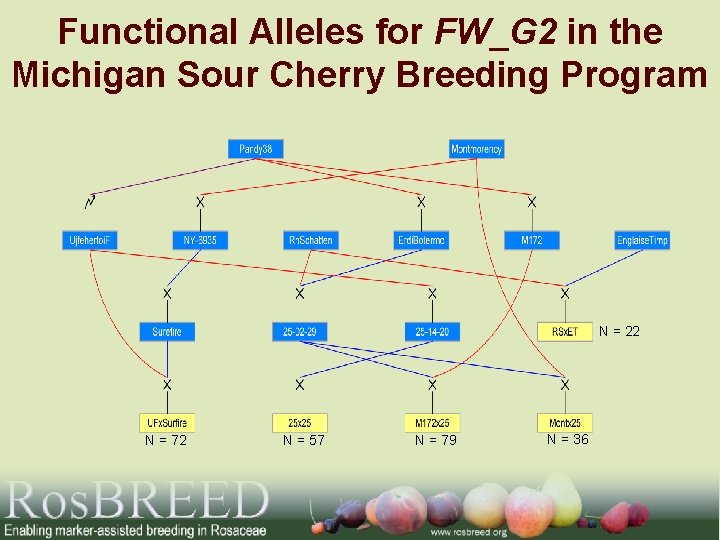
Functional Alleles for FW_G 2 in the Michigan Sour Cherry Breeding Program N = 22 N = 72 N = 57 N = 79 N = 36

Phenotypic data – standardized phenotyping • Standardized phenotyping protocols can be found at http: //www. rosbreed. org/resources/fruit-evaluation

FW_G 2 in tetraploid sour cherry • 17 alleles for the G 2 QTL region were identified in sour cherry using SNP markers • 17 alleles likely an overestimation of the number of functional SNP alleles in the sour cherry breeding materials

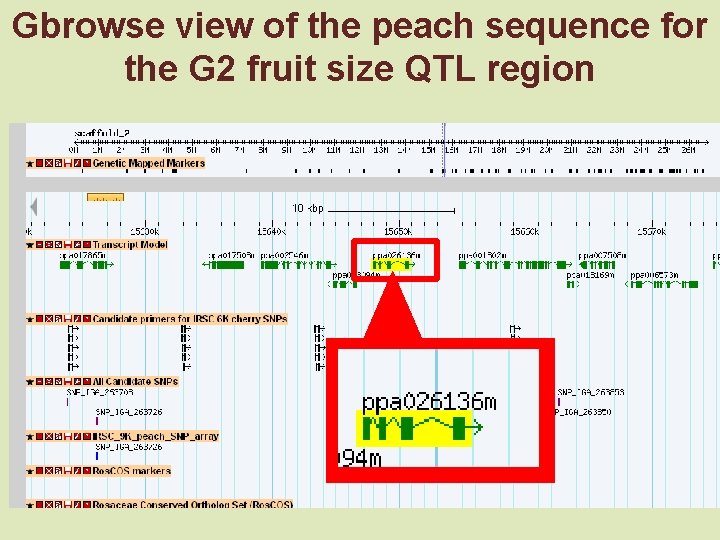
Gbrowse view of the peach sequence for the G 2 fruit size QTL region

Tomato: fw 2. 2 A fruit size gene was discovered in tomato that is a regulator of cell division Tomato and cherry fruit are both enlarged ovaries

Breeding-Friendly DNA Markers Used 3 SSRs • CPSCT 038 • SSR linked to the Cell Number Regulator candidate gene: Pav. CNR 12 & Pce. CNR 12 • BPPCT 034

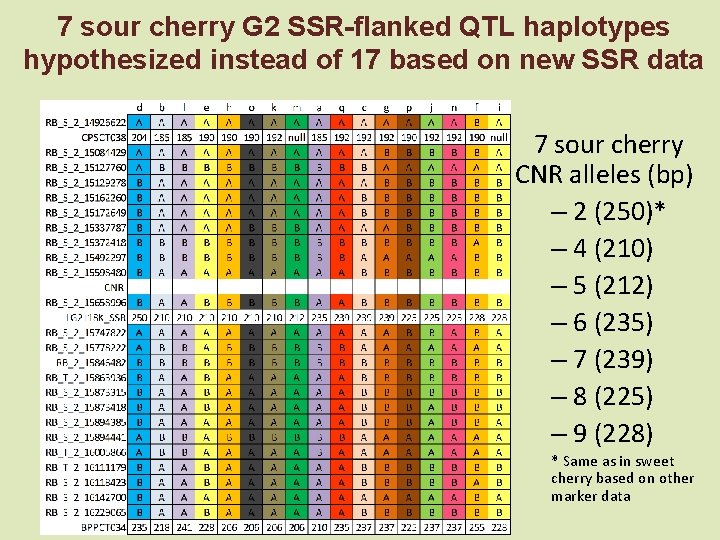
7 sour cherry G 2 SSR-flanked QTL haplotypes hypothesized instead of 17 based on new SSR data 7 sour cherry CNR alleles (bp) – 2 (250)* – 4 (210) – 5 (212) – 6 (235) – 7 (239) – 8 (225) – 9 (228) * Same as in sweet cherry based on other marker data

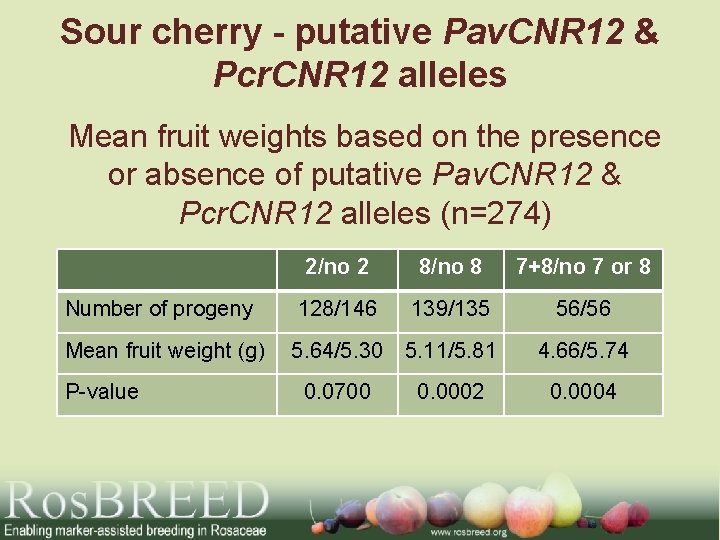
Sour cherry - putative Pav. CNR 12 & Pcr. CNR 12 alleles Mean fruit weights based on the presence or absence of putative Pav. CNR 12 & Pcr. CNR 12 alleles (n=274) 2/no 2 8/no 8 7+8/no 7 or 8 Number of progeny 128/146 139/135 56/56 Mean fruit weight (g) 5. 64/5. 30 5. 11/5. 81 4. 66/5. 74 0. 0700 0. 0002 0. 0004 P-value

FW-G 2 exists in sweet cherry and sour cherry breeding germplasm! • In sweet cherry, 9 ancestral haplotypes for the G 2 region were identified • In sour cherry, 8 ancestral haplotypes for the G 2 region were identified

Other Jewels for Cherry

Other cherry “Jewels” available now! JUN Bloom Time JUL Maturity Date Self-fertility Flesh Color Disease Resistance Fruit Size Firmness Acidity

Conclusion Ros. BREED has and continues to provide DNA tests for valuable traits that have been challenging to plant breeders’ efficiency

Sour cherry breeding program • Prior to Ros. BREED, I had “no clue” about the inheritance of any fruit quality or disease resistance trait in sour cherry. • Now I not only have an understanding of trait inheritance, but I have DNA markers for parent selection, cross design and seedling selection. • The end result is increased breeding efficiency!

Acknowledgements This project is supported by the Specialty Crop Research Initiative of USDA’s National Institute of Food and Agriculture

MSU Amy Iezzoni (PD) Jim Hancock Dechun Wang Cholani Weebadde Univ. of Arkansas John Clark WSU Cameron Peace Dorrie Main Kate Evans Univ. of Minnesota Karina Gallardo Jim Luby Vicki Mc. Cracken Chengyan Yue Nnadozie Oraguzie Former WSU Oregon State Univ. Raymond Jussaume Alexandra Stone Mykel Taylor Cornell Susan Brown Kenong Xu Clemson Ksenija Gasic Gregory Reighard Texas A&M Dave Byrne USDA-ARS Nahla Bassil Gennaro Fazio Univ. of CA-Davis Chad Finn Tom Gradziel Carlos Crisosto Plant Research Intl, Netherlands Univ. of New Hamp. Eric van de Weg Tom Davis Marco Bink
 Qtl club c'est quoi
Qtl club c'est quoi Rf4 inheritance
Rf4 inheritance Qtl mapping
Qtl mapping Qtl moodle
Qtl moodle Harry poter parents
Harry poter parents Jerry wood the outsiders
Jerry wood the outsiders Hardy weinberg equilibrium
Hardy weinberg equilibrium Th
Th Allele frequency def
Allele frequency def Allelic and genotypic frequencies
Allelic and genotypic frequencies Allele vs gene
Allele vs gene Blood type codominance practice problems answers
Blood type codominance practice problems answers How to calculate allele frequency
How to calculate allele frequency Codominance alleles
Codominance alleles Codominance example
Codominance example Hardy weinberg equilibrium
Hardy weinberg equilibrium Allele picture example
Allele picture example Allele vs gene
Allele vs gene Allele vs trait
Allele vs trait What does incomplete dominance mean
What does incomplete dominance mean Allele amorfo
Allele amorfo How to calculate allele frequency from genotype
How to calculate allele frequency from genotype Allèle
Allèle Allele
Allele Codominant
Codominant True allele
True allele True allele
True allele Heterodizomi
Heterodizomi Eye color punnett square
Eye color punnett square Dominant allele definition
Dominant allele definition Test cross definition
Test cross definition Allele genetik
Allele genetik Heredity examples
Heredity examples Allele
Allele Allele neomorfo
Allele neomorfo Allele
Allele Multiple alleles in animals
Multiple alleles in animals Allele frequency
Allele frequency Pku hereditary
Pku hereditary Allele key
Allele key Eye color genetics chart with grandparents
Eye color genetics chart with grandparents Chromosome allele
Chromosome allele Odds ratio
Odds ratio Example of phenotype
Example of phenotype