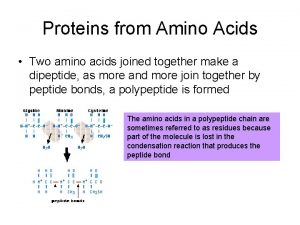
PositionSpecific Weight Matrix Amino acids Position 6 PositionSpecific







- Slides: 24






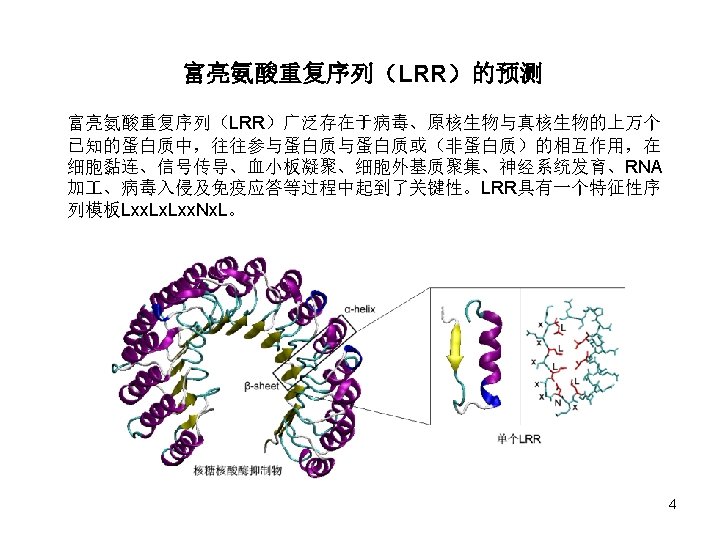
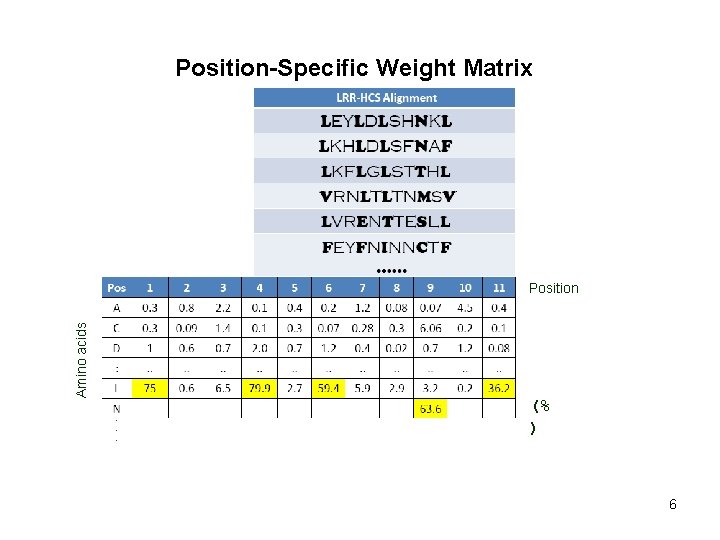
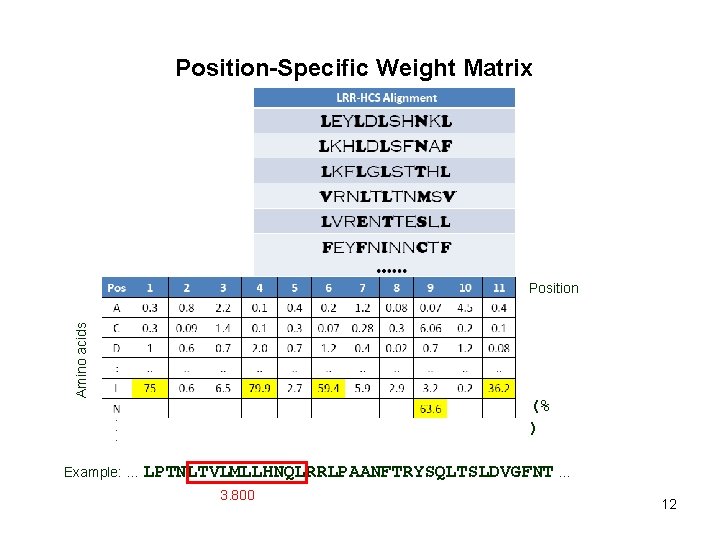
Position-Specific Weight Matrix Amino acids Position (% ) 6

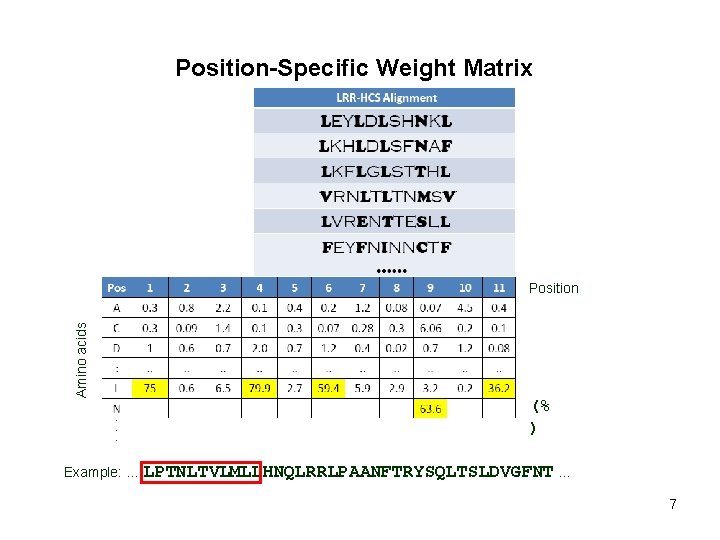
Position-Specific Weight Matrix Amino acids Position (% ) Example: … LPTNLTVLMLLHNQLRRLPAANFTRYSQLTSLDVGFNT … 7

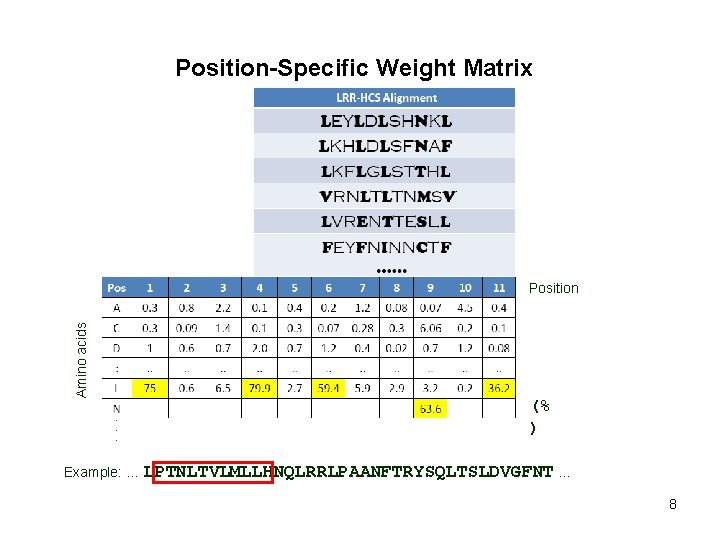
Position-Specific Weight Matrix Amino acids Position (% ) Example: … LPTNLTVLMLLHNQLRRLPAANFTRYSQLTSLDVGFNT … 8

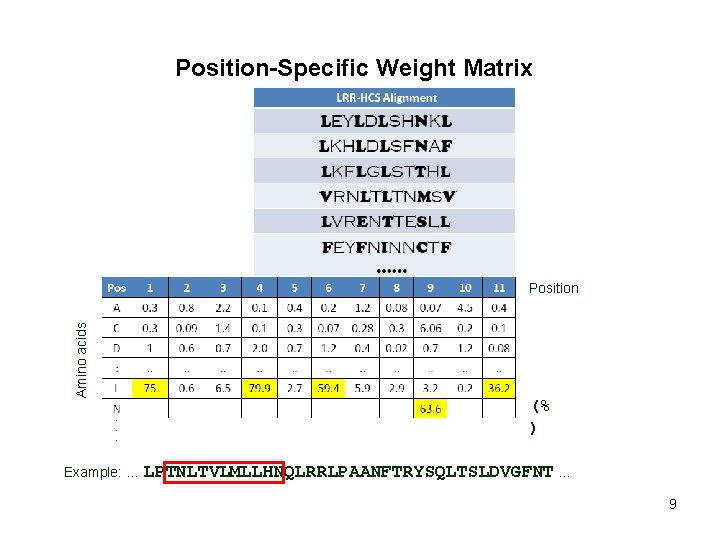
Position-Specific Weight Matrix Amino acids Position (% ) Example: … LPTNLTVLMLLHNQLRRLPAANFTRYSQLTSLDVGFNT … 9

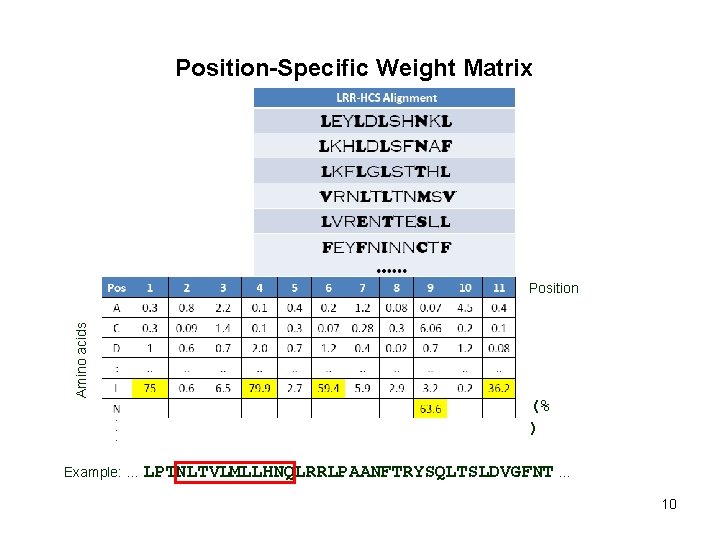
Position-Specific Weight Matrix Amino acids Position (% ) Example: … LPTNLTVLMLLHNQLRRLPAANFTRYSQLTSLDVGFNT … 10

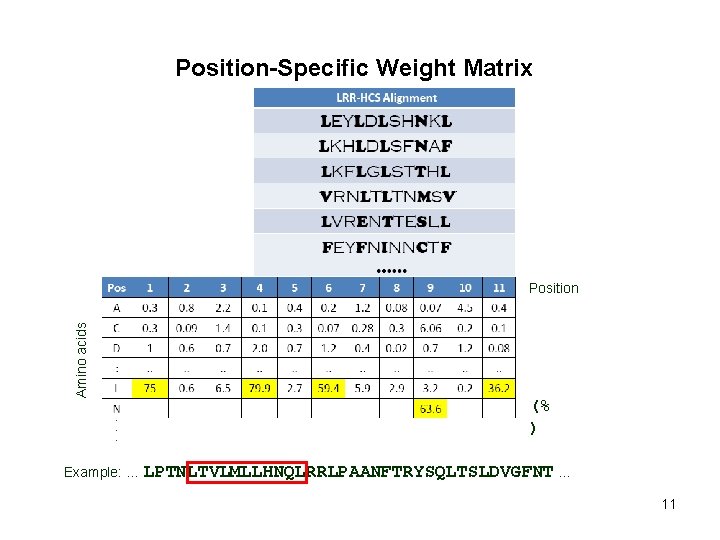
Position-Specific Weight Matrix Amino acids Position (% ) Example: … LPTNLTVLMLLHNQLRRLPAANFTRYSQLTSLDVGFNT … 11

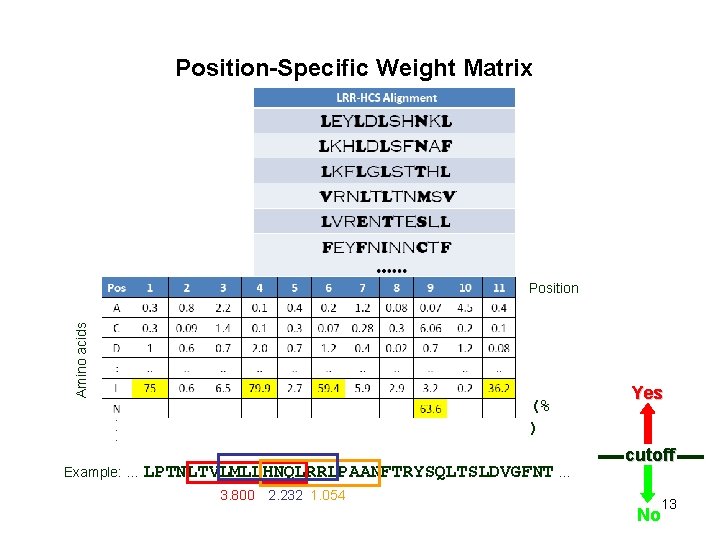
Position-Specific Weight Matrix Amino acids Position (% ) Example: … LPTNLTVLMLLHNQLRRLPAANFTRYSQLTSLDVGFNT … 3. 800 12

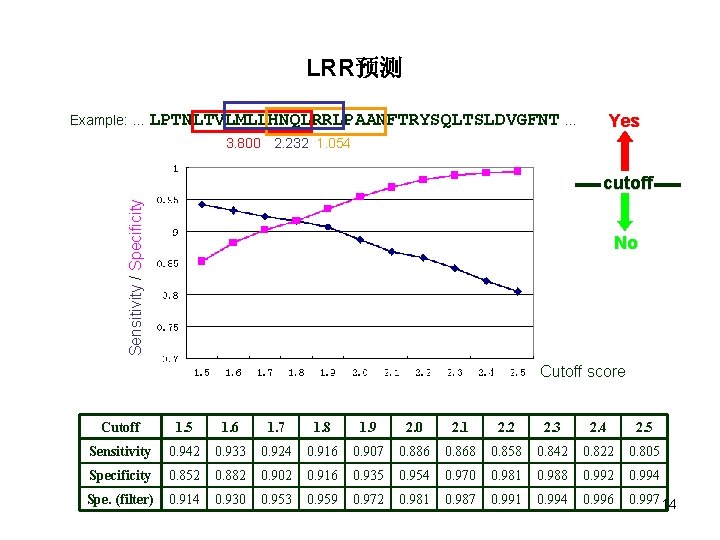
Position-Specific Weight Matrix Amino acids Position (% ) Example: … LPTNLTVLMLLHNQLRRLPAANFTRYSQLTSLDVGFNT … 3. 800 2. 232 1. 054 Yes cutoff 13 No

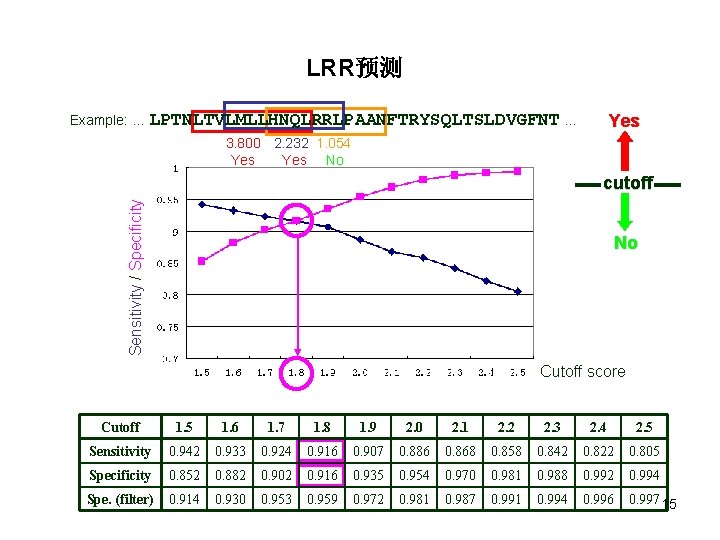
LRR预测 Example: … LPTNLTVLMLLHNQLRRLPAANFTRYSQLTSLDVGFNT … Yes 3. 800 2. 232 1. 054 Sensitivity / Specificity cutoff No Cutoff score Cutoff 1. 5 1. 6 1. 7 1. 8 1. 9 2. 0 2. 1 2. 2 2. 3 2. 4 2. 5 Sensitivity 0. 942 0. 933 0. 924 0. 916 0. 907 0. 886 0. 868 0. 858 0. 842 0. 822 0. 805 Specificity 0. 852 0. 882 0. 902 0. 916 0. 935 0. 954 0. 970 0. 981 0. 988 0. 992 0. 994 Spe. (filter) 0. 914 0. 930 0. 953 0. 959 0. 972 0. 981 0. 987 0. 991 0. 994 0. 996 0. 997 14

LRR预测 Example: … LPTNLTVLMLLHNQLRRLPAANFTRYSQLTSLDVGFNT … Yes 3. 800 2. 232 1. 054 Yes No Sensitivity / Specificity cutoff No Cutoff score Cutoff 1. 5 1. 6 1. 7 1. 8 1. 9 2. 0 2. 1 2. 2 2. 3 2. 4 2. 5 Sensitivity 0. 942 0. 933 0. 924 0. 916 0. 907 0. 886 0. 868 0. 858 0. 842 0. 822 0. 805 Specificity 0. 852 0. 882 0. 902 0. 916 0. 935 0. 954 0. 970 0. 981 0. 988 0. 992 0. 994 Spe. (filter) 0. 914 0. 930 0. 953 0. 959 0. 972 0. 981 0. 987 0. 991 0. 994 0. 996 0. 997 15

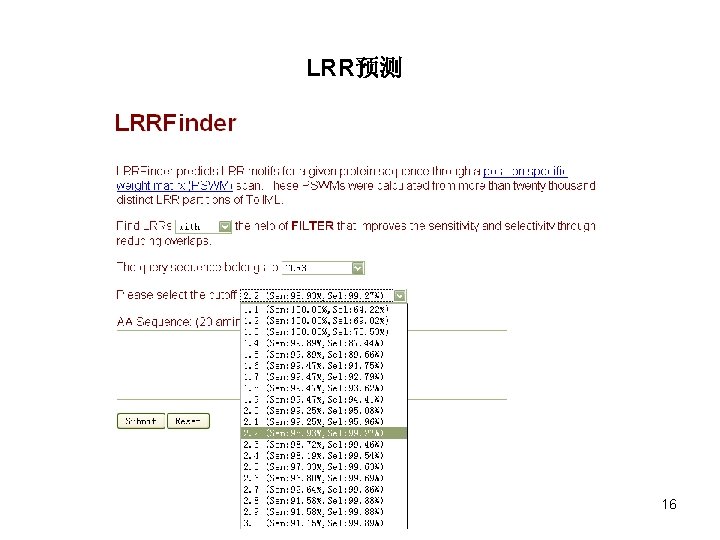
LRR预测 16

LRR预测 17


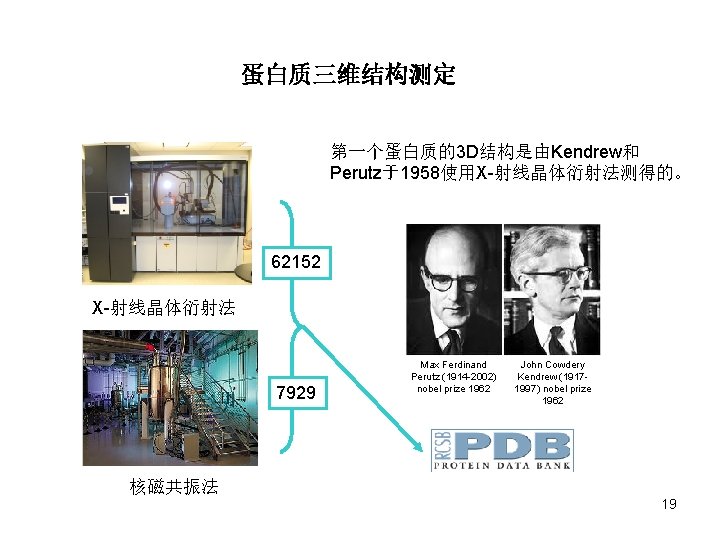
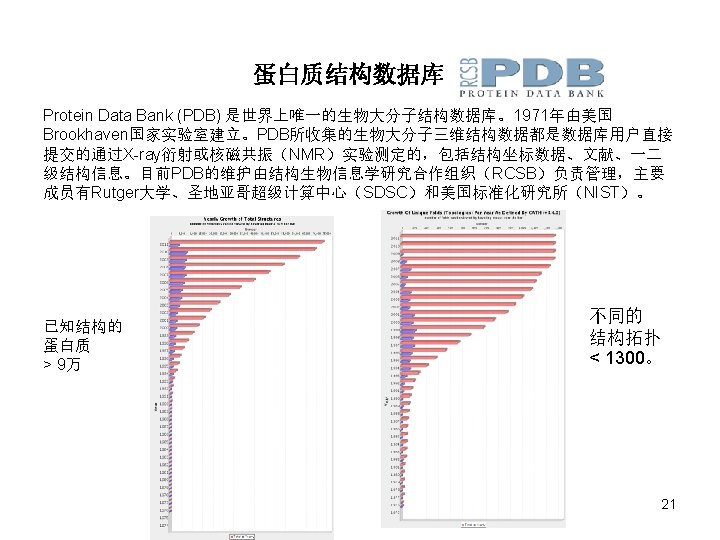
蛋白质三维结构测定 第一个蛋白质的3 D结构是由Kendrew和 Perutz于1958使用X-射线晶体衍射法测得的。 62152 X-射线晶体衍射法 7929 Max Ferdinand Perutz (1914 -2002) nobel prize 1962 John Cowdery Kendrew (19171997) nobel prize 1962 核磁共振法 19





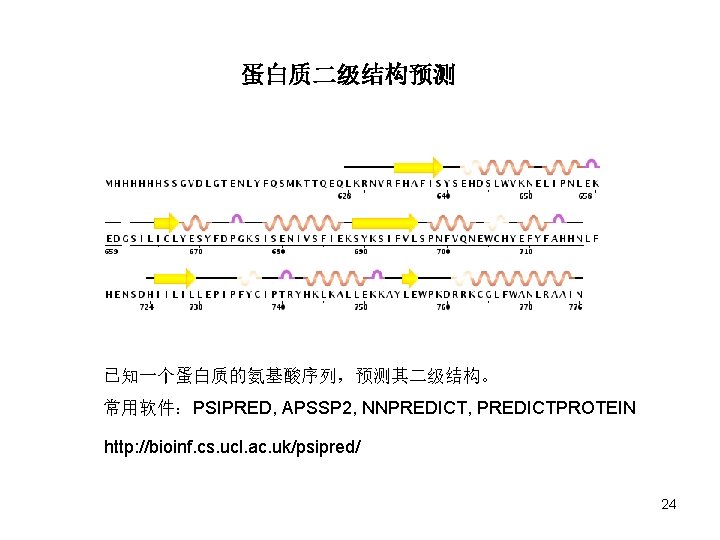
蛋白质二级结构预测 已知一个蛋白质的氨基酸序列,预测其二级结构。 常用软件:PSIPRED, APSSP 2, NNPREDICT, PREDICTPROTEIN http: //bioinf. cs. ucl. ac. uk/psipred/ 24
 Position weight matrix
Position weight matrix Genetic code wheel
Genetic code wheel Amino acids titration curves
Amino acids titration curves Titration curve of amino acids
Titration curve of amino acids Deamination of amino acids
Deamination of amino acids Amino acid condensation reaction
Amino acid condensation reaction Transdeamination of amino acids
Transdeamination of amino acids Amino acids groups
Amino acids groups Early man
Early man Non essential amino acids in food
Non essential amino acids in food Titration curve of amino acids
Titration curve of amino acids Succinly
Succinly Isoleucine ketogenic glucogenic
Isoleucine ketogenic glucogenic Pvt tim hall
Pvt tim hall Amino acid r groups
Amino acid r groups Amino acids classification
Amino acids classification Chemsheets amino acids
Chemsheets amino acids Optical properties of amino acids
Optical properties of amino acids Chymotrypsin cleaves which amino acids
Chymotrypsin cleaves which amino acids Positively charged amino acids
Positively charged amino acids Diphthamide
Diphthamide Non essential amino acids mnemonics
Non essential amino acids mnemonics Dehydration synthesis of amino acids
Dehydration synthesis of amino acids Aromatic amino acids
Aromatic amino acids Phenol containing amino acids
Phenol containing amino acids