Sequence motifs information content logos and Weight matrices











- Slides: 44

Sequence motifs, information content, logos, and Weight matrices • Morten Nielsen, • CBS, Bio. Centrum, • DTU

Objectives • Visualization of binding motifs • Construction of sequence logos • Understand the concepts of weight matrix construction • One of the most important methods of bioinformatics • Realize that sequence profiles are just weight matrices

Outline • Pattern recognition • Regular expressions and probabilities • Information content • Sequence logos • Kullback-Leibler • Multiple alignment and sequence motifs • Weight matrix construction • Sequence weighting • Low (pseudo) counts • Calculate a weight matrix from a sequence alignment • Examples from the real world • HMM’s in immunology • Profile HMMs • MHC class II binding • Gibbs sampling

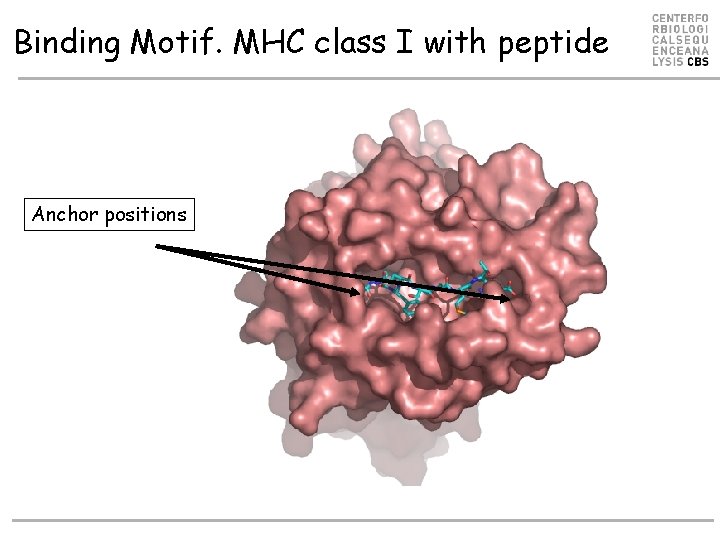
Binding Motif. MHC class I with peptide Anchor positions

Sequence information SLLPAIVEL LLDVPTAAV HLIDYLVTS ILFGHENRV LERPGGNEI PLDGEYFTL ILGFVFTLT KLVALGINA KTWGQYWQV SLLAPGAKQ ILTVILGVL TGAPVTYST GAGIGVAVL KARDPHSGH AVFDRKSDA GLCTLVAML VLHDDLLEA ISNDVCAQV YTAFTIPSI NMFTPYIGV VVLGVVFGI GLYDGMEHL EAAGIGILT YLSTAFARV FLDEFMEGV AAGIGILTV YLLPAIVHI VLFRGGPRG ILAPPVVKL ILMEHIHKL ALSNLEVKL GVLVGVALI LLFGYPVYV DLMGYIPLV TITDQVPFS KIFGSLAFL KVLEYVIKV VIYQYMDDL IAGIGILAI KACDPHSGH LLDFVRFMG FIDSYICQV LMWITQCFL VKTDGNPPE RLMKQDFSV LMIIPLINV ILHNGAYSL KMVELVHFL TLDSQVMSL YLLEMLWRL ALQPGTALL FLPSDFFPS TLWVDPYEV MVDGTLLLL ALFPQLVIL ILDQKINEV ALNELLQHV RTLDKVLEV GLSPTVWLS RLVTLKDIV AFHHVAREL ELVSEFSRM FLWGPRALV VLPDVFIRC LIVIGILIL ACDPHSGHF VLVKSPNHV IISAVVGIL SLLMWITQC SVYDFFVWL RLPRIFCSC TLFIGSHVV MIMVKCWMI YLQLVFGIE STPPPGTRV SLDDYNHLV VLDGLDVLL SVRDRLARL AAGIGILTV GLVPFLVSV YMNGTMSQV GILGFVFTL SLAGGIIGV DLERKVESL HLSTAFARV WLSLLVPFV MLLAVLYCL YLNKIQNSL KLTPLCVTL GLSRYVARL VLPDVFIRC LAGIGLIAA SLYNTVATL GLAPPQHLI VMAGVGSPY QLSLLMWIT FLYGALLLA FLWGPRAYA SLVIVTTFV MLGTHTMEV MLMAQEALA KVAELVHFL RTLDKVLEV SLYSFPEPE SLREWLLRI FLPSDFFPS KLLEPVLLL MLLSVPLLL STNRQSGRQ LLIENVASL FLGENISNF RLDSYVRSL FLPSDFFPS AAGIGILTV MMRKLAILS VLYRYGSFS FLLTRILTI AVGIGIAVV VDGIGILTI RGPGRAFVT LLGRNSFEV LLWTLVVLL LLGATCMFV VLFSSDFRI RLLQETELV VLQWASLAV MLGTHTMEV LMAQEALAF IMIGVLVGV GLPVEYLQV ALYVDSLFF LLSAWILTA AAGIGILTV LLDVPTAAV SLLGLLVEV GLDVLTAKV FLLWATAEA ALSDHHIYL YMNGTMSQV CLGGLLTMV YLEPGPVTA AIMDKNIIL YIGEVLVSV HLGNVKYLV LVVLGLLAV GAGIGVLTA NLVPMVATV PLTFGWCYK SVRDRLARL RLTRFLSRV LMWAKIGPV SLFEGIDFY ILAKFLHWL SLADTNSLA VYDGREHTV ALCRWGLLL KLIANNTRV SLLQHLIGL AAGIGILTV FLWGPRALV LLDVPTAAV ALLPPINIL RILGAVAKV SLPDFGISY GLSEFTEYL GILGFVFTL FIAGNSAYE LLDGTATLR IMDKNIILK CINGVCWTV GIAGGLALL ALGLGLLPV AAGIGIIQI GLHCYEQLV VLEWRFDSR LLMDCSGSI YMDGTMSQV SLLLELEEV SLDQSVVEL STAPPHVNV LLWAARPRL YLSGANLNL LLFAGVQCQ FIYAGSLSA ELTLGEFLK AVPDEIPPL ETVSEQSNV LLDVPTAAV TLIKIQHTL QVCERIPTI KKREEAPSL STAPPAHGV ILKEPVHGV KLGEFYNQM ITDQVPFSV SMVGNWAKV VMNILLQYV GLQDCTMLV GIGIGVLAA QAGIGILLA PLKQHFQIV TLNAWVKVV CLTSTVQLV FLTPKKLQC SLSRFSWGA RLNMFTPYI LLLLTVLTV GVALQTMKQ RMFPNAPYL VLLCESTAV KLVANNTRL MINAYLDKL FAYDGKDYI ITLWQRPLV

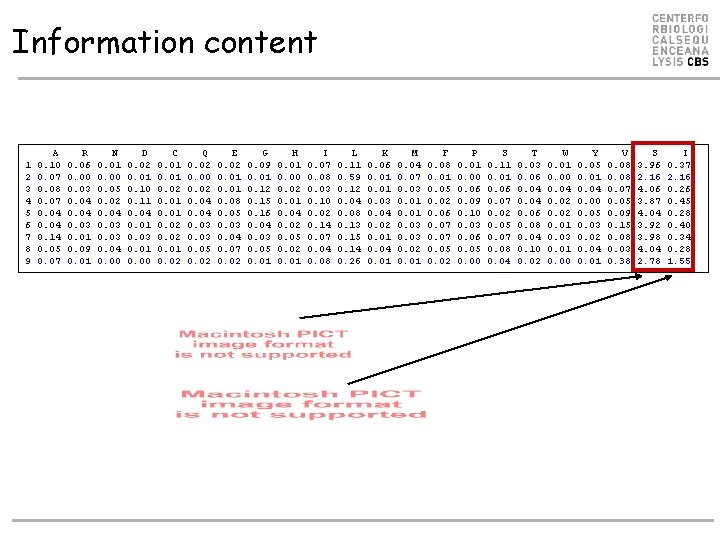
Sequence Information • Say that a peptide must have L at P 2 in order to bind, and that A, F, W, and Y are found at P 1. Which position has most information? • How many questions do I need to ask to tell if a peptide binds looking at only P 1 or P 2? • P 1: 4 questions (at most) • P 2: 1 question (L or not) • P 2 has the most information • Calculate pa at each position • Entropy • Information content • Conserved positions • PV=1, P!v=0 => S=0, I=log(20) • Mutable positions • Paa=1/20 => S=log(20), I=0

Information content 1 2 3 4 5 6 7 8 9 A 0. 10 0. 07 0. 08 0. 07 0. 04 0. 14 0. 05 0. 07 R 0. 06 0. 00 0. 03 0. 04 0. 03 0. 01 0. 09 0. 01 N 0. 01 0. 00 0. 05 0. 02 0. 04 0. 03 0. 04 0. 00 D 0. 02 0. 01 0. 10 0. 11 0. 04 0. 01 0. 03 0. 01 0. 00 C 0. 01 0. 02 Q 0. 02 0. 00 0. 02 0. 04 0. 03 0. 05 0. 02 E 0. 02 0. 01 0. 08 0. 05 0. 03 0. 04 0. 07 0. 02 G 0. 09 0. 01 0. 12 0. 15 0. 16 0. 04 0. 03 0. 05 0. 01 H 0. 01 0. 00 0. 02 0. 01 0. 04 0. 02 0. 05 0. 02 0. 01 I 0. 07 0. 08 0. 03 0. 10 0. 02 0. 14 0. 07 0. 04 0. 08 L 0. 11 0. 59 0. 12 0. 04 0. 08 0. 13 0. 15 0. 14 0. 26 K 0. 06 0. 01 0. 03 0. 04 0. 02 0. 01 0. 04 0. 01 M 0. 04 0. 07 0. 03 0. 01 0. 03 0. 02 0. 01 F 0. 08 0. 01 0. 05 0. 02 0. 06 0. 07 0. 05 0. 02 P 0. 01 0. 00 0. 06 0. 09 0. 10 0. 03 0. 06 0. 05 0. 00 S 0. 11 0. 06 0. 07 0. 02 0. 05 0. 07 0. 08 0. 04 T 0. 03 0. 06 0. 04 0. 06 0. 08 0. 04 0. 10 0. 02 W 0. 01 0. 00 0. 04 0. 02 0. 01 0. 03 0. 01 0. 00 Y 0. 05 0. 01 0. 04 0. 00 0. 05 0. 03 0. 02 0. 04 0. 01 V 0. 08 0. 07 0. 05 0. 09 0. 15 0. 08 0. 03 0. 38 S 3. 96 2. 16 4. 06 3. 87 4. 04 3. 92 3. 98 4. 04 2. 78 I 0. 37 2. 16 0. 26 0. 45 0. 28 0. 40 0. 34 0. 28 1. 55

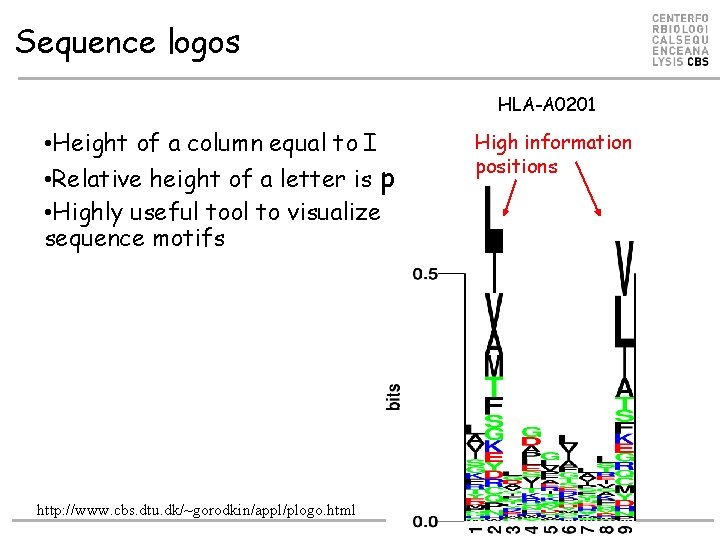
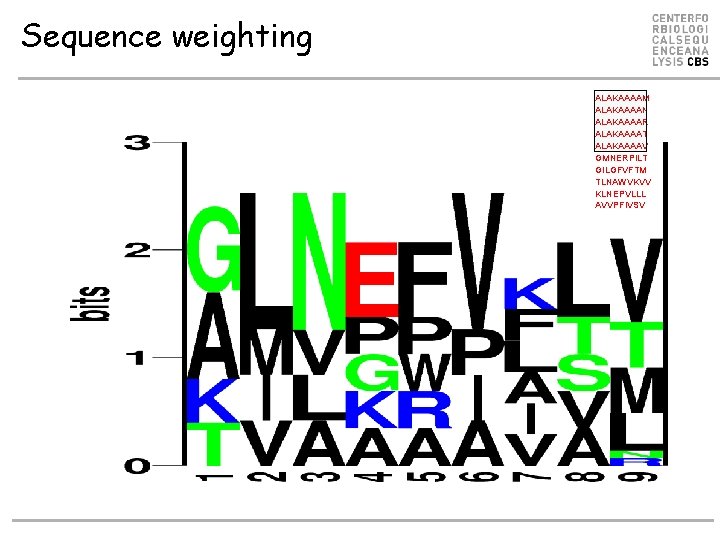
Sequence logos HLA-A 0201 • Height of a column equal to I • Relative height of a letter is p • Highly useful tool to visualize sequence motifs http: //www. cbs. dtu. dk/~gorodkin/appl/plogo. html High information positions

Kullback Leibler Logo

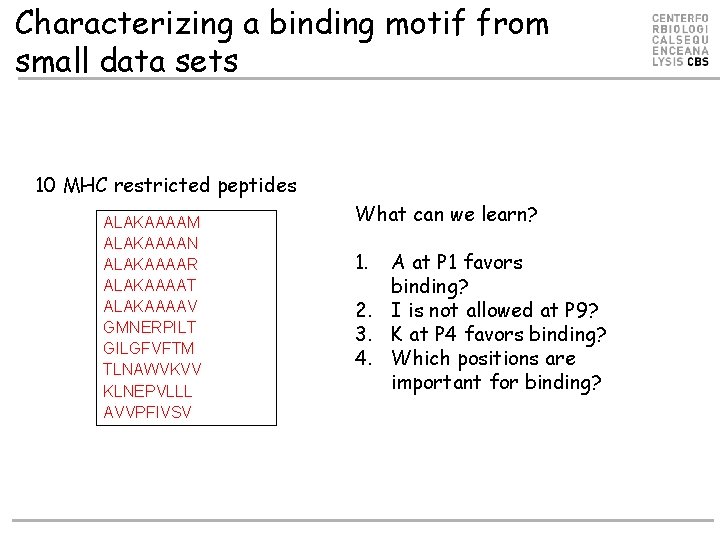
Characterizing a binding motif from small data sets 10 MHC restricted peptides ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV What can we learn? 1. A at P 1 favors binding? 2. I is not allowed at P 9? 3. K at P 4 favors binding? 4. Which positions are important for binding?

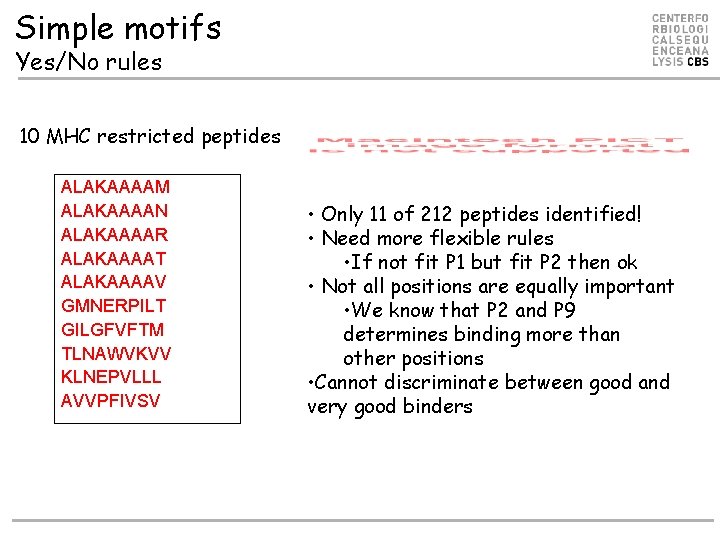
Simple motifs Yes/No rules 10 MHC restricted peptides ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV • Only 11 of 212 peptides identified! • Need more flexible rules • If not fit P 1 but fit P 2 then ok • Not all positions are equally important • We know that P 2 and P 9 determines binding more than other positions • Cannot discriminate between good and very good binders

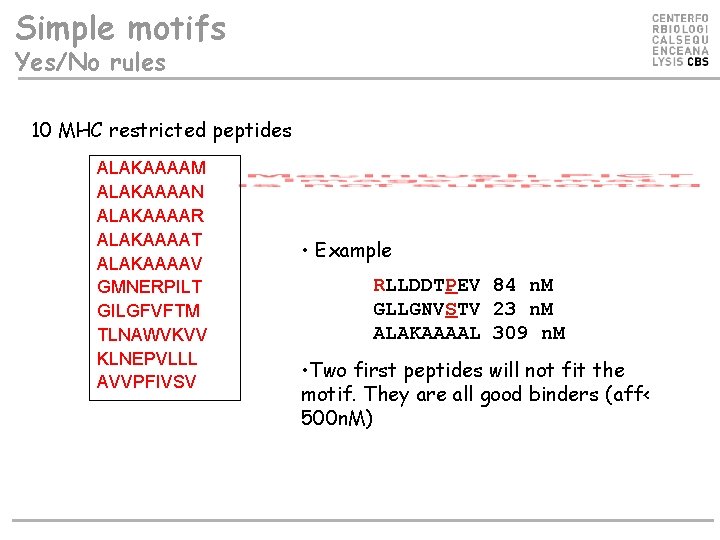
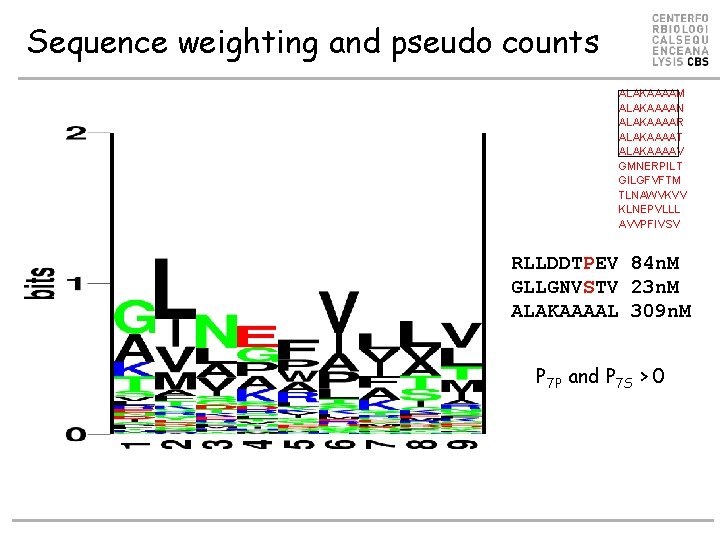
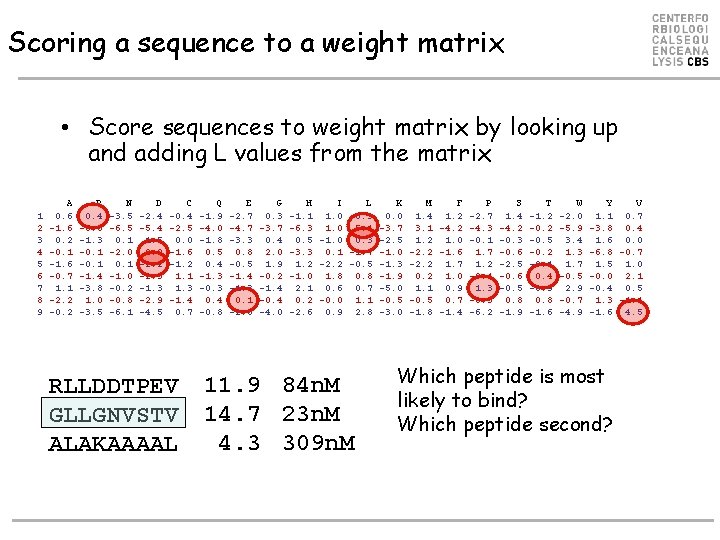
Simple motifs Yes/No rules 10 MHC restricted peptides ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV • Example RLLDDTPEV 84 n. M GLLGNVSTV 23 n. M ALAKAAAAL 309 n. M • Two first peptides will not fit the motif. They are all good binders (aff< 500 n. M)

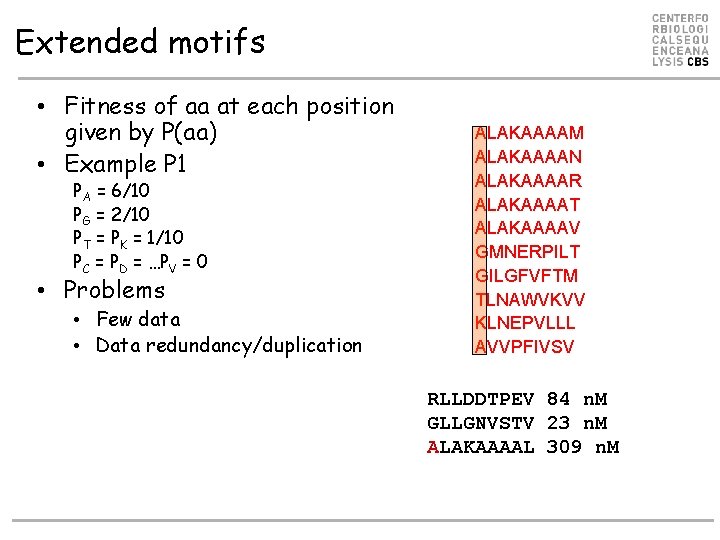
Extended motifs • Fitness of aa at each position given by P(aa) • Example P 1 PA = 6/10 PG = 2/10 PT = PK = 1/10 PC = PD = …PV = 0 • Problems • Few data • Data redundancy/duplication ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV RLLDDTPEV 84 n. M GLLGNVSTV 23 n. M ALAKAAAAL 309 n. M

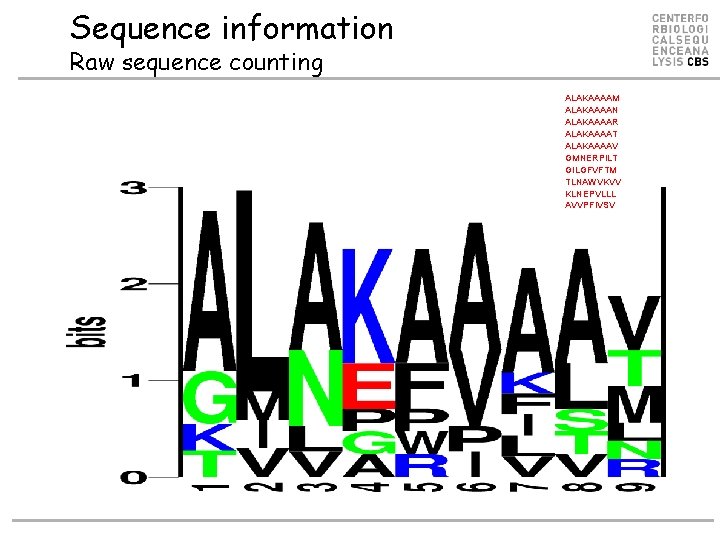
Sequence information Raw sequence counting ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV

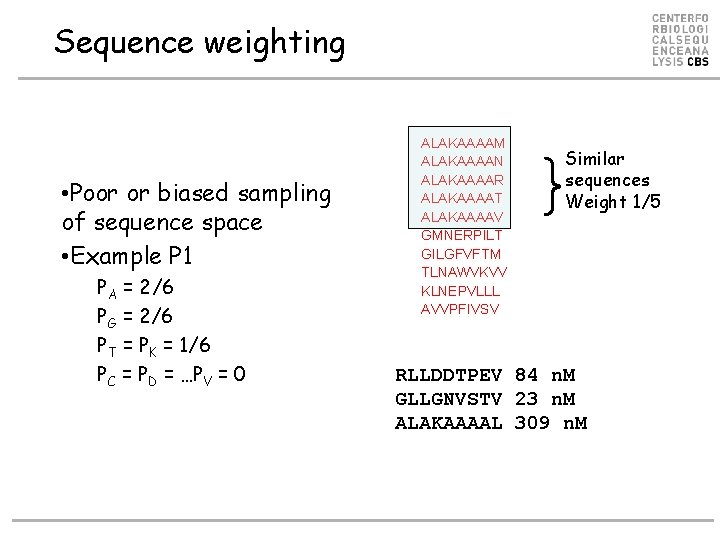
Sequence weighting • Poor or biased sampling of sequence space • Example P 1 PA = 2/6 PG = 2/6 PT = PK = 1/6 PC = PD = …PV = 0 ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV } Similar sequences Weight 1/5 RLLDDTPEV 84 n. M GLLGNVSTV 23 n. M ALAKAAAAL 309 n. M

Sequence weighting ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV

Pseudo counts • I is not found at position P 9. Does this mean that I is forbidden (P(I)=0)? • No! Use Blosum substitution matrix to estimate pseudo frequency of I at P 9 ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV

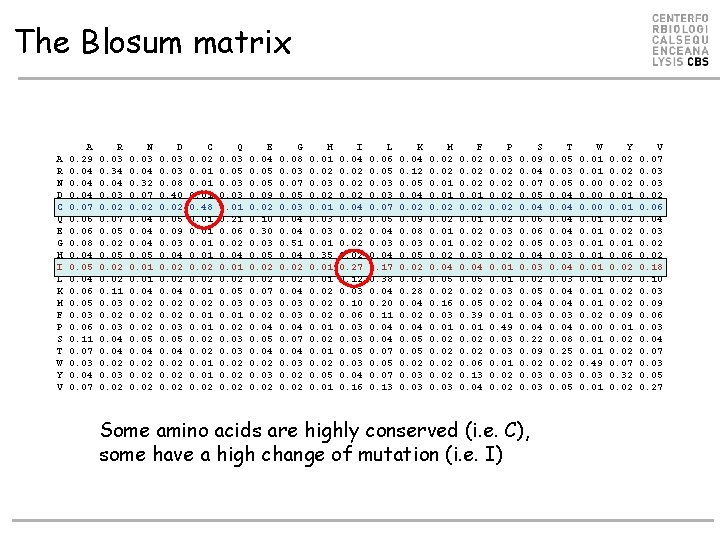
The Blosum matrix A R N D C Q E G H I L K M F P S T W Y V A 0. 29 0. 04 0. 07 0. 06 0. 08 0. 04 0. 05 0. 04 0. 06 0. 05 0. 03 0. 06 0. 11 0. 07 0. 03 0. 04 0. 07 R 0. 03 0. 34 0. 03 0. 02 0. 07 0. 05 0. 02 0. 11 0. 03 0. 02 0. 03 0. 04 0. 02 0. 03 0. 02 N 0. 03 0. 04 0. 32 0. 07 0. 02 0. 04 0. 05 0. 01 0. 04 0. 02 0. 05 0. 04 0. 02 D 0. 03 0. 08 0. 40 0. 02 0. 05 0. 09 0. 03 0. 04 0. 02 0. 03 0. 05 0. 04 0. 02 C 0. 02 0. 01 0. 48 0. 01 0. 02 0. 01 0. 02 Q 0. 03 0. 05 0. 03 0. 01 0. 21 0. 06 0. 02 0. 04 0. 01 0. 02 0. 05 0. 03 0. 01 0. 02 0. 03 0. 02 E 0. 04 0. 05 0. 09 0. 02 0. 10 0. 30 0. 03 0. 05 0. 02 0. 07 0. 03 0. 02 0. 04 0. 05 0. 04 0. 02 0. 03 0. 02 G 0. 08 0. 03 0. 07 0. 05 0. 03 0. 04 0. 51 0. 04 0. 02 0. 04 0. 03 0. 04 0. 07 0. 04 0. 03 0. 02 H 0. 01 0. 02 0. 03 0. 02 0. 01 0. 03 0. 01 0. 35 0. 01 0. 02 0. 05 0. 01 I 0. 04 0. 02 0. 04 0. 03 0. 02 0. 27 0. 12 0. 03 0. 10 0. 06 0. 03 0. 05 0. 03 0. 04 0. 16 L 0. 06 0. 05 0. 03 0. 07 0. 05 0. 04 0. 03 0. 04 0. 17 0. 38 0. 04 0. 20 0. 11 0. 04 0. 07 0. 05 0. 07 0. 13 K 0. 04 0. 12 0. 05 0. 04 0. 02 0. 09 0. 08 0. 03 0. 05 0. 02 0. 03 0. 28 0. 04 0. 02 0. 04 0. 05 0. 02 0. 03 M 0. 02 0. 01 0. 02 0. 04 0. 05 0. 02 0. 16 0. 03 0. 01 0. 02 0. 03 F 0. 02 0. 01 0. 02 0. 03 0. 04 0. 05 0. 02 0. 05 0. 39 0. 01 0. 02 0. 06 0. 13 0. 04 P 0. 03 0. 02 0. 01 0. 49 0. 03 0. 01 0. 02 S 0. 09 0. 04 0. 07 0. 05 0. 04 0. 06 0. 05 0. 04 0. 03 0. 02 0. 05 0. 04 0. 03 0. 04 0. 22 0. 09 0. 02 0. 03 Some amino acids are highly conserved (i. e. C), some have a high change of mutation (i. e. I) T 0. 05 0. 03 0. 05 0. 04 0. 03 0. 04 0. 08 0. 25 0. 02 0. 03 0. 05 W 0. 01 0. 00 0. 01 0. 02 0. 00 0. 01 0. 49 0. 03 0. 01 Y 0. 02 0. 01 0. 06 0. 02 0. 09 0. 01 0. 02 0. 07 0. 32 0. 02 V 0. 07 0. 03 0. 02 0. 06 0. 04 0. 03 0. 02 0. 18 0. 10 0. 03 0. 09 0. 06 0. 03 0. 04 0. 07 0. 03 0. 05 0. 27

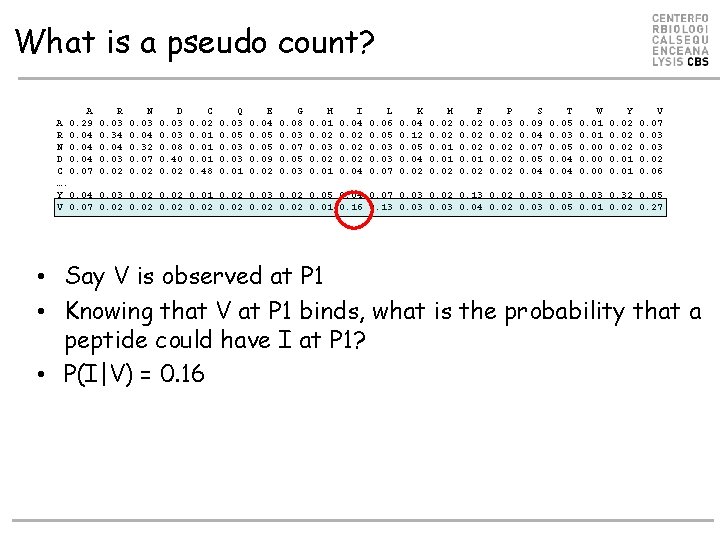
What is a pseudo count? A A 0. 29 R 0. 04 N 0. 04 D 0. 04 C 0. 07 …. Y 0. 04 V 0. 07 R 0. 03 0. 34 0. 03 0. 02 N 0. 03 0. 04 0. 32 0. 07 0. 02 D 0. 03 0. 08 0. 40 0. 02 C 0. 02 0. 01 0. 48 Q 0. 03 0. 05 0. 03 0. 01 E 0. 04 0. 05 0. 09 0. 02 G 0. 08 0. 03 0. 07 0. 05 0. 03 H 0. 01 0. 02 0. 03 0. 02 0. 01 I 0. 04 0. 02 0. 04 L 0. 06 0. 05 0. 03 0. 07 K 0. 04 0. 12 0. 05 0. 04 0. 02 M 0. 02 0. 01 0. 02 F 0. 02 0. 01 0. 02 P 0. 03 0. 02 S 0. 09 0. 04 0. 07 0. 05 0. 04 T 0. 05 0. 03 0. 05 0. 04 W 0. 01 0. 00 Y 0. 02 0. 01 V 0. 07 0. 03 0. 02 0. 06 0. 03 0. 02 0. 01 0. 02 0. 03 0. 02 0. 05 0. 04 0. 07 0. 03 0. 02 0. 13 0. 02 0. 03 0. 32 0. 05 0. 02 0. 01 0. 16 0. 13 0. 03 0. 04 0. 02 0. 03 0. 05 0. 01 0. 02 0. 27 • Say V is observed at P 1 • Knowing that V at P 1 binds, what is the probability that a peptide could have I at P 1? • P(I|V) = 0. 16

Pseudo count estimation • Calculate observed amino acids frequencies fa • Pseudo frequency for amino acid b • Example ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV

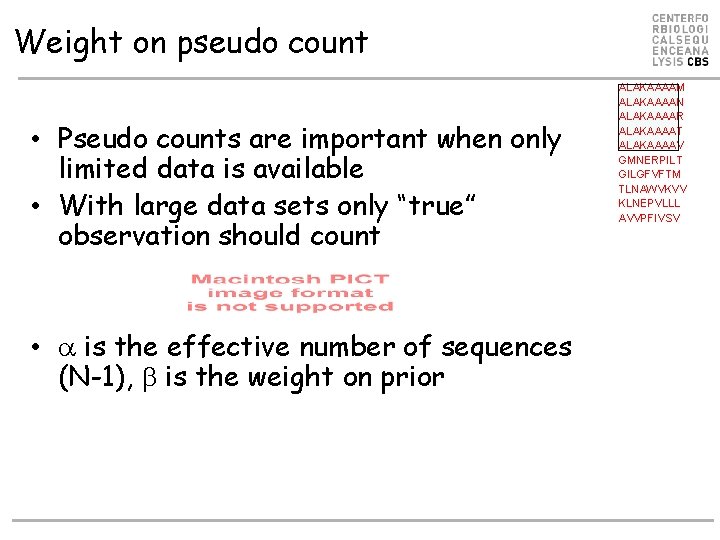
Weight on pseudo count • Pseudo counts are important when only limited data is available • With large data sets only “true” observation should count • is the effective number of sequences (N-1), is the weight on prior ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV

Weight on pseudo count • Example • If large, p ≈ f and only the observed data defines the motif • If small, p ≈ g and the pseudo counts (or prior) defines the motif • is [50 -200] normally ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV

Sequence weighting and pseudo counts ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV RLLDDTPEV 84 n. M GLLGNVSTV 23 n. M ALAKAAAAL 309 n. M P 7 P and P 7 S > 0

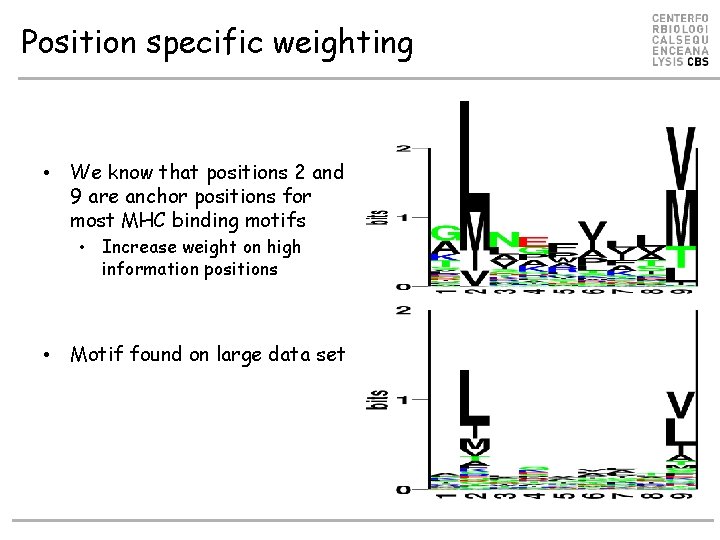
Position specific weighting • We know that positions 2 and 9 are anchor positions for most MHC binding motifs • Increase weight on high information positions • Motif found on large data set

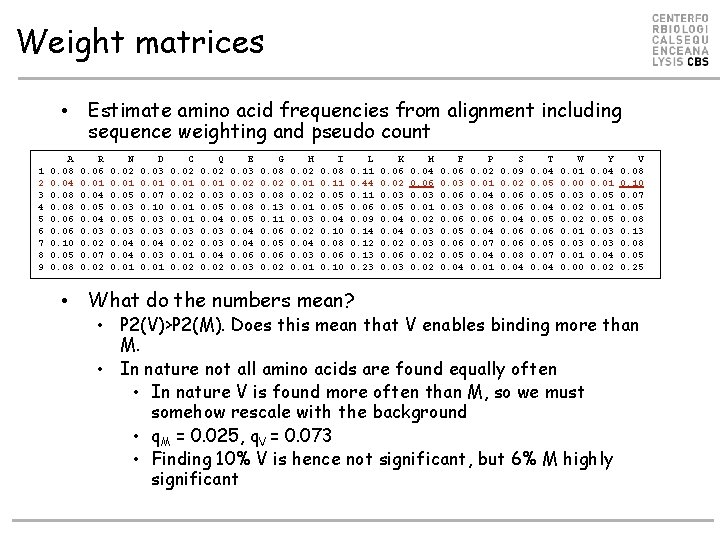
Weight matrices • Estimate amino acid frequencies from alignment including sequence weighting and pseudo count 1 2 3 4 5 6 7 8 9 A 0. 08 0. 04 0. 08 0. 06 0. 10 0. 05 0. 08 R 0. 06 0. 01 0. 04 0. 05 0. 04 0. 03 0. 02 0. 07 0. 02 N 0. 02 0. 01 0. 05 0. 03 0. 04 0. 01 D 0. 03 0. 01 0. 07 0. 10 0. 03 0. 04 0. 03 0. 01 C 0. 02 0. 01 0. 03 0. 02 0. 01 0. 02 Q 0. 02 0. 01 0. 03 0. 05 0. 04 0. 03 0. 04 0. 02 E 0. 03 0. 02 0. 03 0. 08 0. 05 0. 04 0. 06 0. 03 G 0. 08 0. 02 0. 08 0. 13 0. 11 0. 06 0. 05 0. 06 0. 02 H 0. 02 0. 01 0. 03 0. 02 0. 04 0. 03 0. 01 I 0. 08 0. 11 0. 05 0. 04 0. 10 0. 08 0. 06 0. 10 L 0. 11 0. 44 0. 11 0. 06 0. 09 0. 14 0. 12 0. 13 0. 23 • What do the numbers mean? K 0. 06 0. 02 0. 03 0. 05 0. 04 0. 02 0. 06 0. 03 M 0. 04 0. 06 0. 03 0. 01 0. 02 0. 03 0. 02 F 0. 06 0. 03 0. 06 0. 05 0. 04 P 0. 02 0. 01 0. 04 0. 08 0. 06 0. 04 0. 07 0. 04 0. 01 S 0. 09 0. 02 0. 06 0. 04 0. 06 0. 08 0. 04 T 0. 04 0. 05 0. 06 0. 05 0. 07 0. 04 W 0. 01 0. 00 0. 03 0. 02 0. 01 0. 03 0. 01 0. 00 Y 0. 04 0. 01 0. 05 0. 03 0. 04 0. 02 V 0. 08 0. 10 0. 07 0. 05 0. 08 0. 13 0. 08 0. 05 0. 25 • P 2(V)>P 2(M). Does this mean that V enables binding more than M. • In nature not all amino acids are found equally often • In nature V is found more often than M, so we must somehow rescale with the background • q. M = 0. 025, q. V = 0. 073 • Finding 10% V is hence not significant, but 6% M highly significant

Weight matrices • A weight matrix is given as Wij = log(pij/qj) • where i is a position in the motif, and j an amino acid. qj is the background frequency for amino acid j. 1 2 3 4 5 6 7 8 9 A 0. 6 -1. 6 0. 2 -0. 1 -1. 6 -0. 7 1. 1 -2. 2 -0. 2 R 0. 4 -6. 6 -1. 3 -0. 1 -1. 4 -3. 8 1. 0 -3. 5 N -3. 5 -6. 5 0. 1 -2. 0 0. 1 -1. 0 -0. 2 -0. 8 -6. 1 D -2. 4 -5. 4 1. 5 2. 0 -2. 2 -2. 3 -1. 3 -2. 9 -4. 5 C -0. 4 -2. 5 0. 0 -1. 6 -1. 2 1. 1 1. 3 -1. 4 0. 7 Q -1. 9 -4. 0 -1. 8 0. 5 0. 4 -1. 3 -0. 3 0. 4 -0. 8 E -2. 7 -4. 7 -3. 3 0. 8 -0. 5 -1. 4 -1. 3 0. 1 -2. 5 G 0. 3 -3. 7 0. 4 2. 0 1. 9 -0. 2 -1. 4 -0. 4 -4. 0 H I L K M F -1. 1 1. 0 0. 3 0. 0 1. 4 1. 2 -6. 3 1. 0 5. 1 -3. 7 3. 1 -4. 2 0. 5 -1. 0 0. 3 -2. 5 1. 2 1. 0 -3. 3 0. 1 -1. 7 -1. 0 -2. 2 -1. 6 1. 2 -2. 2 -0. 5 -1. 3 -2. 2 1. 7 -1. 0 1. 8 0. 8 -1. 9 0. 2 1. 0 2. 1 0. 6 0. 7 -5. 0 1. 1 0. 9 0. 2 -0. 0 1. 1 -0. 5 0. 7 -2. 6 0. 9 2. 8 -3. 0 -1. 8 -1. 4 • W is a L x 20 matrix, L is motif length P -2. 7 -4. 3 -0. 1 1. 7 1. 2 -0. 4 1. 3 -0. 3 -6. 2 S 1. 4 -4. 2 -0. 3 -0. 6 -2. 5 -0. 6 -0. 5 0. 8 -1. 9 T -1. 2 -0. 5 -0. 2 -0. 1 0. 4 -0. 9 0. 8 -1. 6 W -2. 0 -5. 9 3. 4 1. 3 1. 7 -0. 5 2. 9 -0. 7 -4. 9 Y V 1. 1 0. 7 -3. 8 0. 4 1. 6 0. 0 -6. 8 -0. 7 1. 5 1. 0 -0. 0 2. 1 -0. 4 0. 5 1. 3 -1. 1 -1. 6 4. 5

Scoring a sequence to a weight matrix • Score sequences to weight matrix by looking up and adding L values from the matrix 1 2 3 4 5 6 7 8 9 A 0. 6 -1. 6 0. 2 -0. 1 -1. 6 -0. 7 1. 1 -2. 2 -0. 2 R 0. 4 -6. 6 -1. 3 -0. 1 -1. 4 -3. 8 1. 0 -3. 5 N -3. 5 -6. 5 0. 1 -2. 0 0. 1 -1. 0 -0. 2 -0. 8 -6. 1 D -2. 4 -5. 4 1. 5 2. 0 -2. 2 -2. 3 -1. 3 -2. 9 -4. 5 C -0. 4 -2. 5 0. 0 -1. 6 -1. 2 1. 1 1. 3 -1. 4 0. 7 RLLDDTPEV GLLGNVSTV ALAKAAAAL Q -1. 9 -4. 0 -1. 8 0. 5 0. 4 -1. 3 -0. 3 0. 4 -0. 8 E -2. 7 -4. 7 -3. 3 0. 8 -0. 5 -1. 4 -1. 3 0. 1 -2. 5 G 0. 3 -3. 7 0. 4 2. 0 1. 9 -0. 2 -1. 4 -0. 4 -4. 0 H I L K M F -1. 1 1. 0 0. 3 0. 0 1. 4 1. 2 -6. 3 1. 0 5. 1 -3. 7 3. 1 -4. 2 0. 5 -1. 0 0. 3 -2. 5 1. 2 1. 0 -3. 3 0. 1 -1. 7 -1. 0 -2. 2 -1. 6 1. 2 -2. 2 -0. 5 -1. 3 -2. 2 1. 7 -1. 0 1. 8 0. 8 -1. 9 0. 2 1. 0 2. 1 0. 6 0. 7 -5. 0 1. 1 0. 9 0. 2 -0. 0 1. 1 -0. 5 0. 7 -2. 6 0. 9 2. 8 -3. 0 -1. 8 -1. 4 11. 9 84 n. M 14. 7 23 n. M 4. 3 309 n. M P -2. 7 -4. 3 -0. 1 1. 7 1. 2 -0. 4 1. 3 -0. 3 -6. 2 S 1. 4 -4. 2 -0. 3 -0. 6 -2. 5 -0. 6 -0. 5 0. 8 -1. 9 T -1. 2 -0. 5 -0. 2 -0. 1 0. 4 -0. 9 0. 8 -1. 6 W -2. 0 -5. 9 3. 4 1. 3 1. 7 -0. 5 2. 9 -0. 7 -4. 9 Y V 1. 1 0. 7 -3. 8 0. 4 1. 6 0. 0 -6. 8 -0. 7 1. 5 1. 0 -0. 0 2. 1 -0. 4 0. 5 1. 3 -1. 1 -1. 6 4. 5 Which peptide is most likely to bind? Which peptide second?

An example!! (See handout)

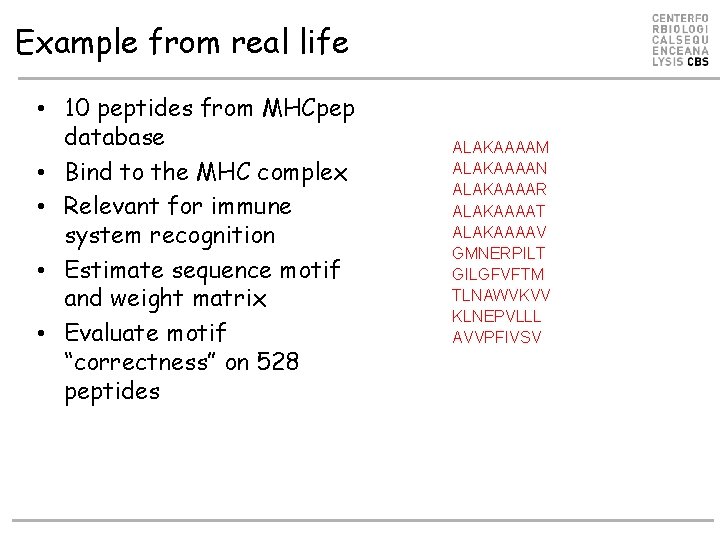
Example from real life • 10 peptides from MHCpep database • Bind to the MHC complex • Relevant for immune system recognition • Estimate sequence motif and weight matrix • Evaluate motif “correctness” on 528 peptides ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV

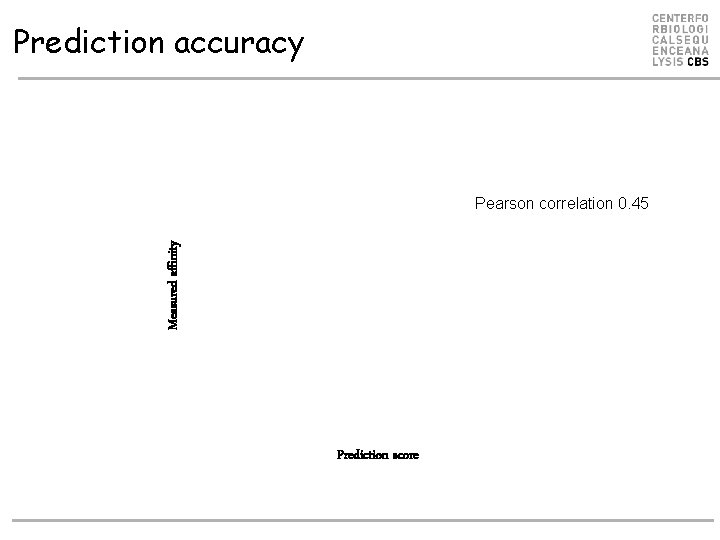
Prediction accuracy Measured affinity Pearson correlation 0. 45 Prediction score

Predictive performance

End of first part • Take a deep breath • Smile to you neighbor

Hidden Markov Models • Weight matrices do not deal with insertions and deletions • In alignments, this is done in an ad-hoc manner by optimization of the two gap penalties for first gap and gap extension • HMM is a natural frame work where insertions/deletions are dealt with explicitly

Profile HMM’s • Alignments based on conventional scoring matrices (BLOSUM 62) scores all positions in a sequence in an equal manner • Some positions are highly conserved, some are highly variable (more than what is described in the BLOSUM matrix) • Profile HMM’s are ideal suited to describe such position specific variations

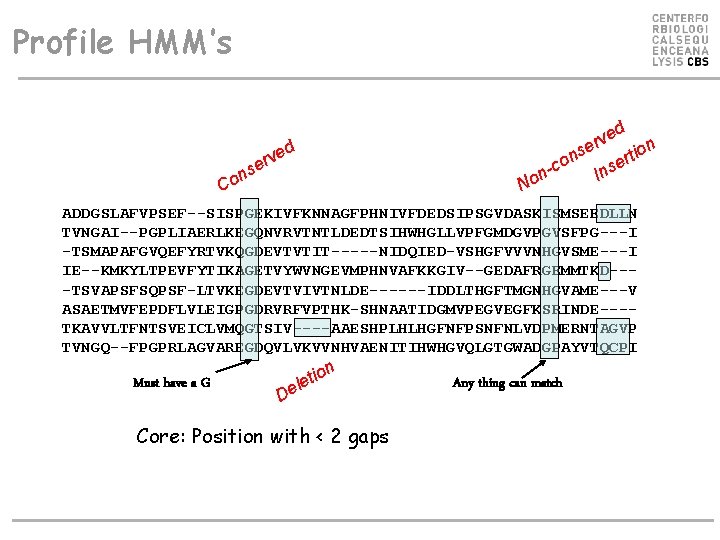
Profile HMM’s ed v r on se i t n r o e n-c Ins d e v r e ns o C No ADDGSLAFVPSEF--SISPGEKIVFKNNAGFPHNIVFDEDSIPSGVDASKISMSEEDLLN TVNGAI--PGPLIAERLKEGQNVRVTNTLDEDTSIHWHGLLVPFGMDGVPGVSFPG---I -TSMAPAFGVQEFYRTVKQGDEVTVTIT-----NIDQIED-VSHGFVVVNHGVSME---I IE--KMKYLTPEVFYTIKAGETVYWVNGEVMPHNVAFKKGIV--GEDAFRGEMMTKD---TSVAPSFSQPSF-LTVKEGDEVTVIVTNLDE------IDDLTHGFTMGNHGVAME---V ASAETMVFEPDFLVLEIGPGDRVRFVPTHK-SHNAATIDGMVPEGVEGFKSRINDE---TKAVVLTFNTSVEICLVMQGTSIV----AAESHPLHLHGFNFPSNFNLVDPMERNTAGVP TVNGQ--FPGPRLAGVAREGDQVLVKVVNHVAENITIHWHGVQLGTGWADGPAYVTQCPI Must have a G on i t le De Core: Position with < 2 gaps Any thing can match

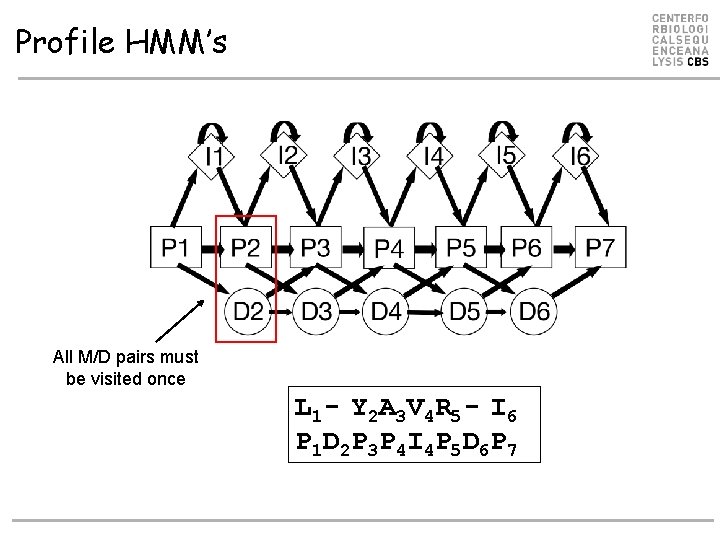
Profile HMM’s All M/D pairs must be visited once L 1 - Y 2 A 3 V 4 R 5 - I 6 P 1 D 2 P 3 P 4 I 4 P 5 D 6 P 7

HMM’s and weight matrices • To achieve high performance, the emission frequencies are estimated using the techniques of • Sequence weighting • Pseudo counts • In the case of un-gapped alignments HMM’s become simple weight matrices

HMM vs. alignment • Detailed description of core • Conserved/variable positions • Price for insertions/deletions varies at different locations in sequence • These features cannot be captured in conventional alignments

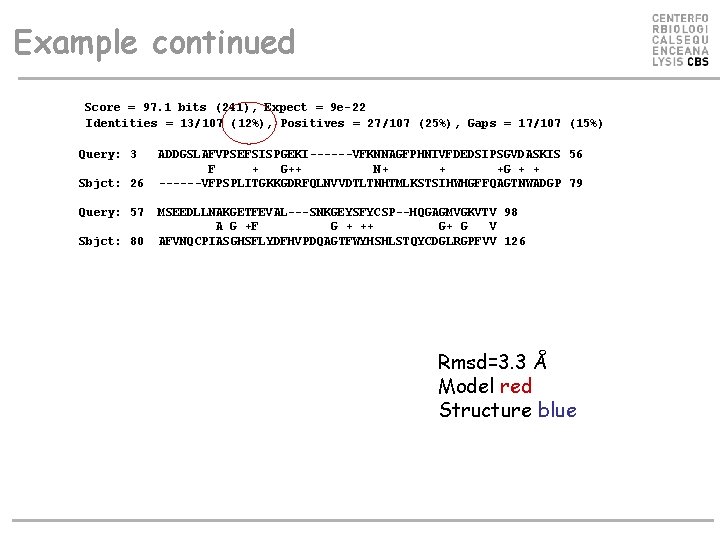
Example. Sequence profiles • Alignment of protein sequences 1 PLC. _ and 1 GYC. A • E-value > 1000 • Profile alignment • Align 1 PLC. _ against Swiss-prot • Make position specific weight matrix from alignment • Use this matrix to align 1 PLC. _ against 1 GYC. A • E-value < 10 -22. Rmsd=3. 3

Example continued Score = 97. 1 bits (241), Expect = 9 e-22 Identities = 13/107 (12%), Positives = 27/107 (25%), Gaps = 17/107 (15%) Query: 3 Sbjct: 26 Query: 57 Sbjct: 80 ADDGSLAFVPSEFSISPGEKI------VFKNNAGFPHNIVFDEDSIPSGVDASKIS 56 F + G++ N+ + +G + + ------VFPSPLITGKKGDRFQLNVVDTLTNHTMLKSTSIHWHGFFQAGTNWADGP 79 MSEEDLLNAKGETFEVAL---SNKGEYSFYCSP--HQGAGMVGKVTV 98 A G +F G + ++ G+ G V AFVNQCPIASGHSFLYDFHVPDQAGTFWYHSHLSTQYCDGLRGPFVV 126 Rmsd=3. 3 Å Model red Structure blue

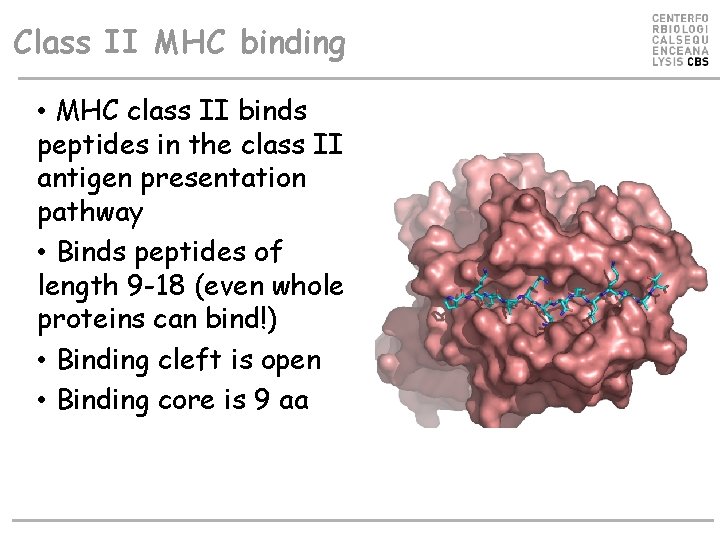
Class II MHC binding • MHC class II binds peptides in the class II antigen presentation pathway • Binds peptides of length 9 -18 (even whole proteins can bind!) • Binding cleft is open • Binding core is 9 aa

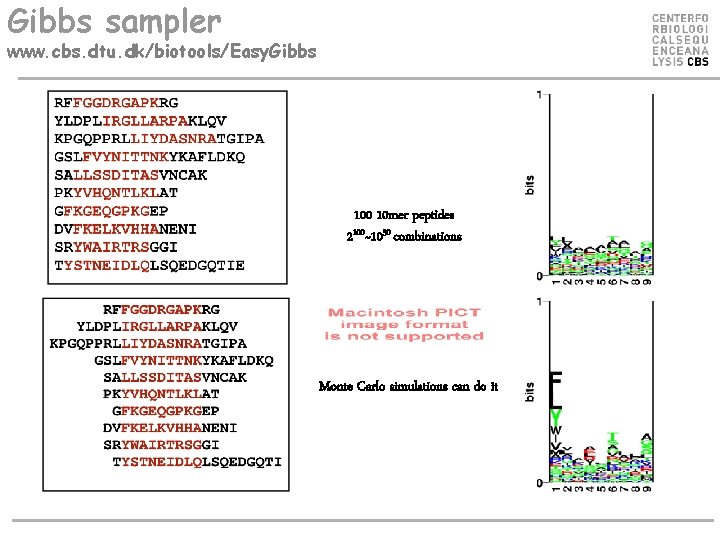
Gibbs sampler www. cbs. dtu. dk/biotools/Easy. Gibbs 100 10 mer peptides 2100~1030 combinations Monte Carlo simulations can do it

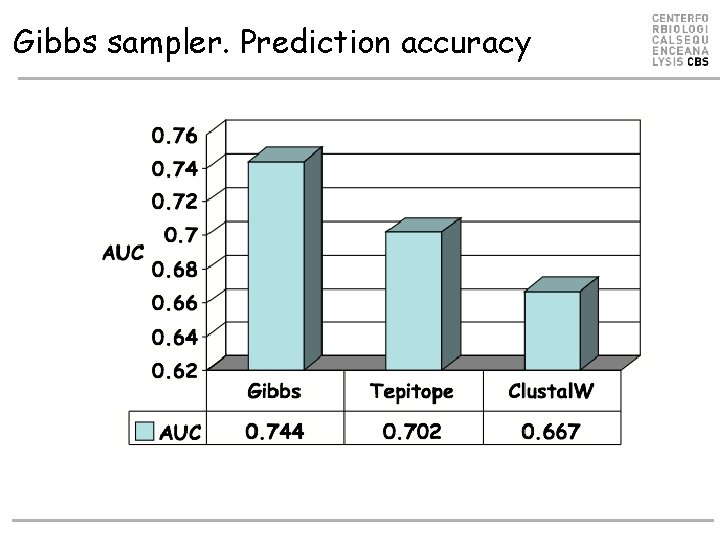
Gibbs sampler. Prediction accuracy

Summary • Weight matrices can accurately describe a sequence motif like MHC class I • Use sequence weighting to remove data redundancy • Use pseudo count to compensate for few data points • HMMs are specialized weight matrices • Sequence profiles are gapless HMM’s • Gibbs sampling can detect MHC class II binding motif