Sequence motifs information content logos and HMMs Morten
![Mutual information P 1 I(i, j) = Saai Saaj P(aai, aaj) * log[P(aai, aaj)/P(aai)*P(aaj)] Mutual information P 1 I(i, j) = Saai Saaj P(aai, aaj) * log[P(aai, aaj)/P(aai)*P(aaj)]](https://slidetodoc.com/presentation_image_h2/b073db05fba5248a1f1455d0d80fe454/image-12.jpg)





- Slides: 29

Sequence motifs, information content, logos, and HMM’s Morten Nielsen, CBS, Bio. Centrum, DTU

Outline l l l Multiple alignments and sequence motifs Weight matrices and consensus sequence – Sequence weighting – Low (pseudo) counts Information content – Sequence logos – Mutual information Example from the real world HMM’s and profile HMM’s – TMHMM (trans-membrane protein) – Gene finding Links to HMM packages

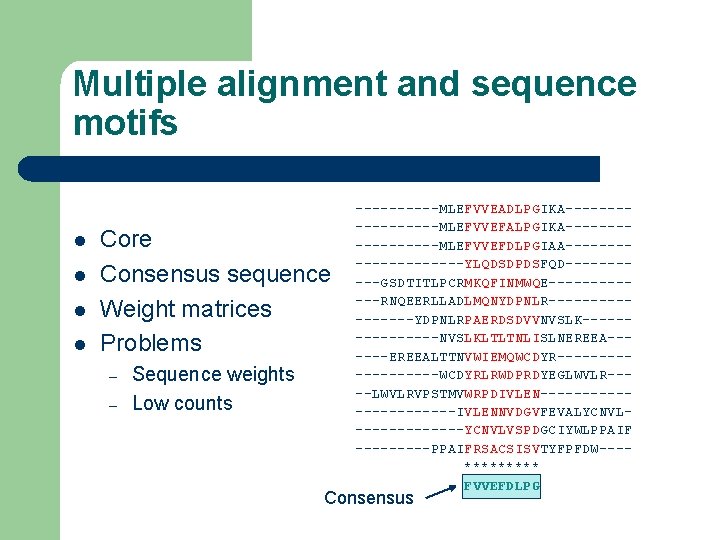
Multiple alignment and sequence motifs l l Core Consensus sequence Weight matrices Problems – – Sequence weights Low counts -----MLEFVVEADLPGIKA---------MLEFVVEFALPGIKA---------MLEFVVEFDLPGIAA----------YLQDSDPDSFQD-----GSDTITLPCRMKQFINMWQE------RNQEERLLADLMQNYDPNLR--------YDPNLRPAERDSDVVNVSLK--------NVSLKLTLTNLISLNEREEA------EREEALTTNVWIEMQWCDYR---------WCDYRLRWDPRDYEGLWVLR----LWVLRVPSTMVWRPDIVLEN-----------IVLENNVDGVFEVALYCNVL-------YCNVLVSPDGCIYWLPPAIF -----PPAIFRSACSISVTYFPFDW---***** FVVEFDLPG Consensus

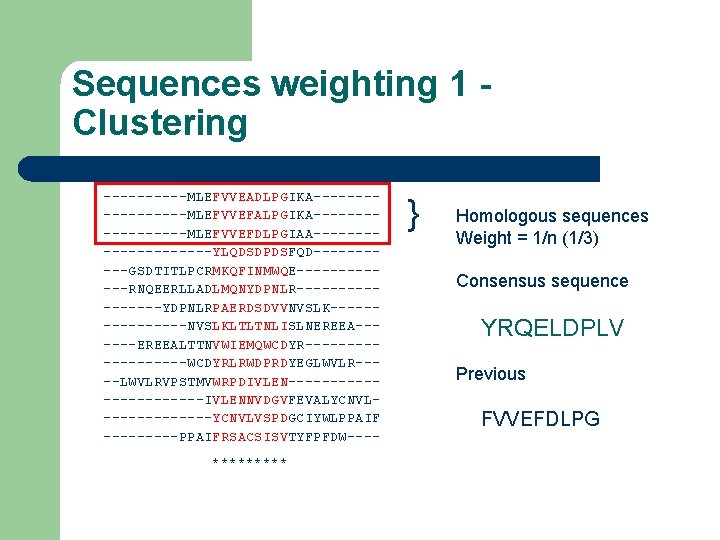
Sequences weighting 1 Clustering -----MLEFVVEADLPGIKA---------MLEFVVEFALPGIKA---------MLEFVVEFDLPGIAA----------YLQDSDPDSFQD-----GSDTITLPCRMKQFINMWQE------RNQEERLLADLMQNYDPNLR--------YDPNLRPAERDSDVVNVSLK--------NVSLKLTLTNLISLNEREEA------EREEALTTNVWIEMQWCDYR---------WCDYRLRWDPRDYEGLWVLR----LWVLRVPSTMVWRPDIVLEN-----------IVLENNVDGVFEVALYCNVL-------YCNVLVSPDGCIYWLPPAIF -----PPAIFRSACSISVTYFPFDW---***** } Homologous sequences Weight = 1/n (1/3) Consensus sequence YRQELDPLV Previous FVVEFDLPG

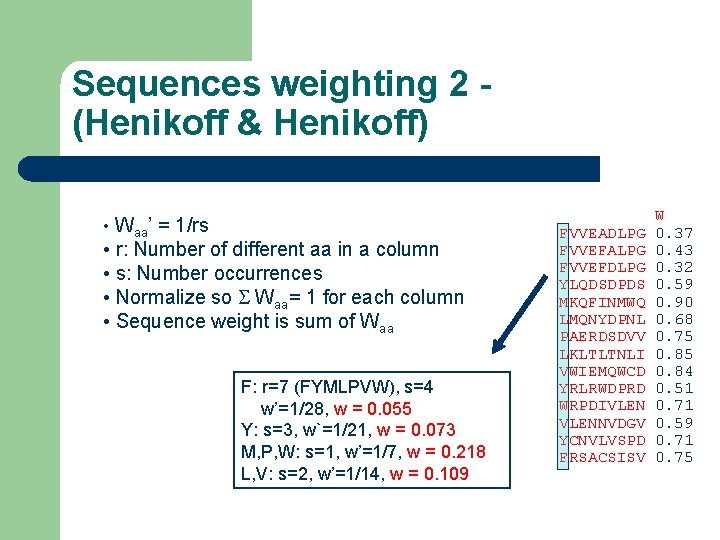
Sequences weighting 2 (Henikoff & Henikoff) • Waa’ = 1/rs • r: Number of different aa in a column • s: Number occurrences • Normalize so S Waa= 1 for each column • Sequence weight is sum of Waa F: r=7 (FYMLPVW), s=4 w’=1/28, w = 0. 055 Y: s=3, w`=1/21, w = 0. 073 M, P, W: s=1, w’=1/7, w = 0. 218 L, V: s=2, w’=1/14, w = 0. 109 FVVEADLPG FVVEFALPG FVVEFDLPG YLQDSDPDS MKQFINMWQ LMQNYDPNL PAERDSDVV LKLTLTNLI VWIEMQWCD YRLRWDPRD WRPDIVLENNVDGV YCNVLVSPD FRSACSISV W 0. 37 0. 43 0. 32 0. 59 0. 90 0. 68 0. 75 0. 84 0. 51 0. 71 0. 59 0. 71 0. 75

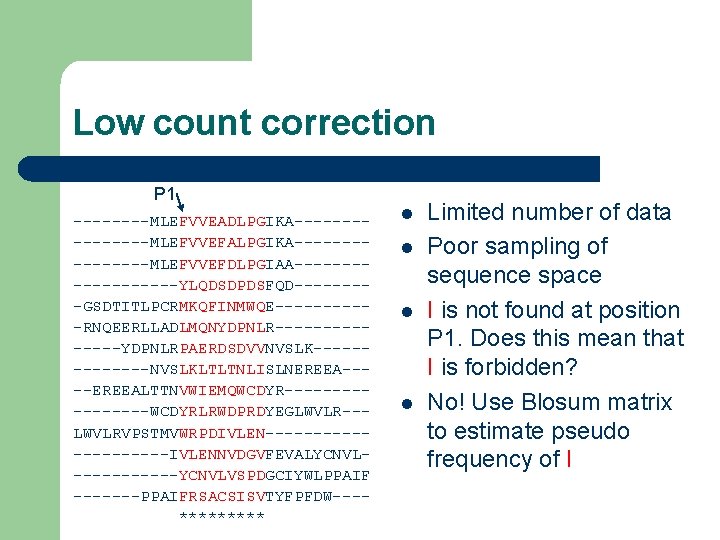
Low count correction P 1 ----MLEFVVEADLPGIKA--------MLEFVVEFALPGIKA--------MLEFVVEFDLPGIAA---------YLQDSDPDSFQD----GSDTITLPCRMKQFINMWQE-----RNQEERLLADLMQNYDPNLR-------YDPNLRPAERDSDVVNVSLK-------NVSLKLTLTNLISLNEREEA----EREEALTTNVWIEMQWCDYR--------WCDYRLRWDPRDYEGLWVLR--LWVLRVPSTMVWRPDIVLEN----------IVLENNVDGVFEVALYCNVL------YCNVLVSPDGCIYWLPPAIF -------PPAIFRSACSISVTYFPFDW---***** l l Limited number of data Poor sampling of sequence space I is not found at position P 1. Does this mean that I is forbidden? No! Use Blosum matrix to estimate pseudo frequency of I

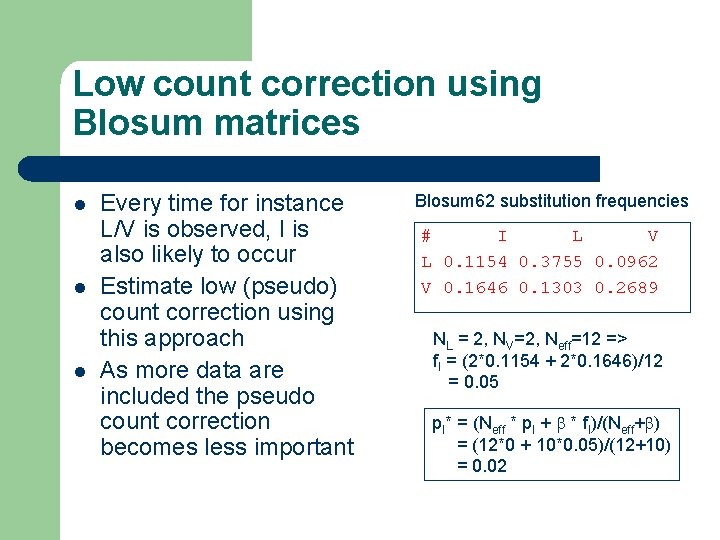
Low count correction using Blosum matrices l l l Every time for instance L/V is observed, I is also likely to occur Estimate low (pseudo) count correction using this approach As more data are included the pseudo count correction becomes less important Blosum 62 substitution frequencies # I L V L 0. 1154 0. 3755 0. 0962 V 0. 1646 0. 1303 0. 2689 NL = 2, NV=2, Neff=12 => f. I = (2*0. 1154 + 2*0. 1646)/12 = 0. 05 p. I* = (Neff * p. I + b * f. I)/(Neff+b) = (12*0 + 10*0. 05)/(12+10) = 0. 02

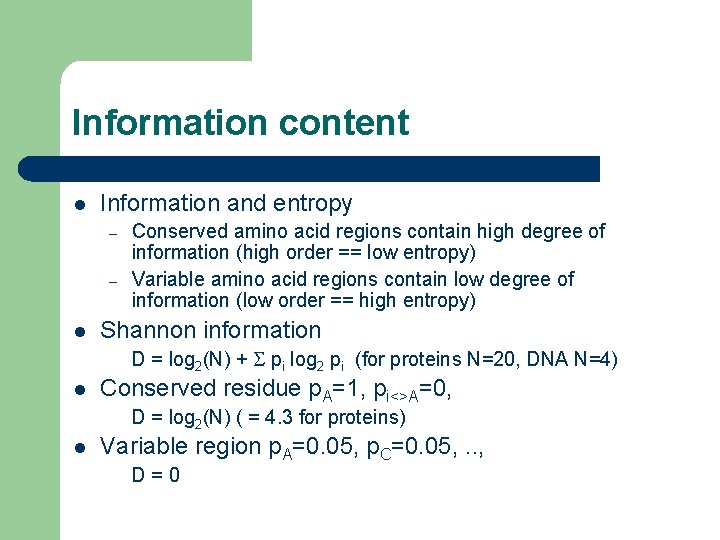
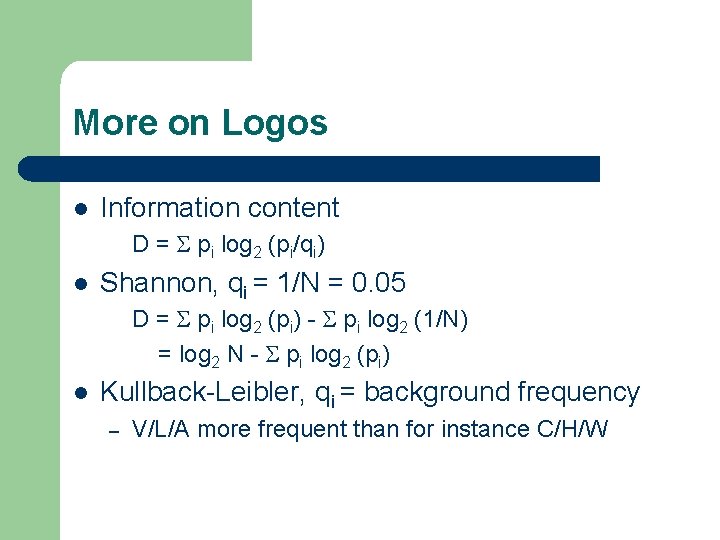
Information content l Information and entropy – – l Conserved amino acid regions contain high degree of information (high order == low entropy) Variable amino acid regions contain low degree of information (low order == high entropy) Shannon information D = log 2(N) + S pi log 2 pi (for proteins N=20, DNA N=4) l Conserved residue p. A=1, pi<>A=0, D = log 2(N) ( = 4. 3 for proteins) l Variable region p. A=0. 05, p. C=0. 05, . . , D=0

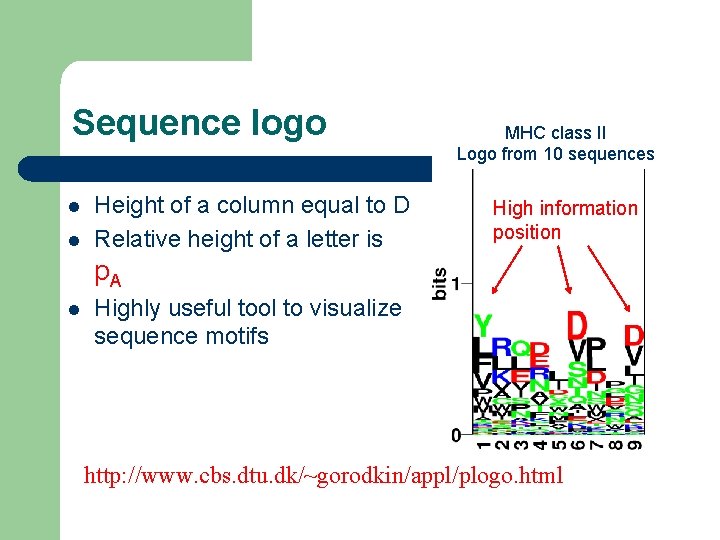
Sequence logo l l Height of a column equal to D Relative height of a letter is MHC class II Logo from 10 sequences High information position p. A l Highly useful tool to visualize sequence motifs http: //www. cbs. dtu. dk/~gorodkin/appl/plogo. html

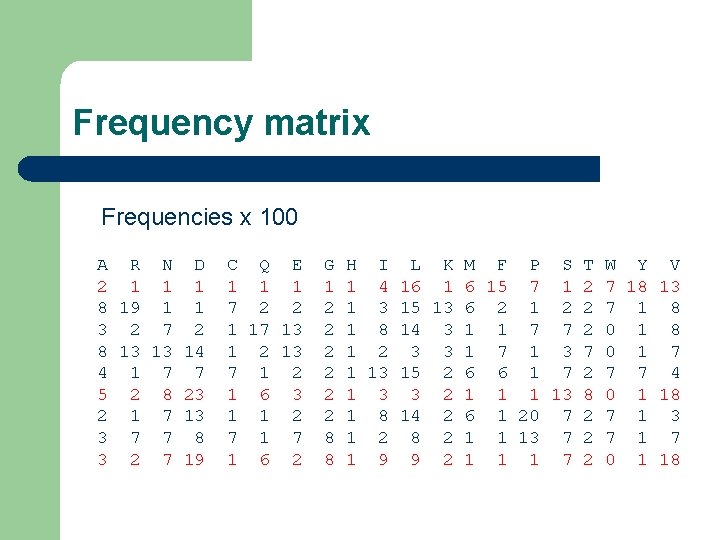
Frequency matrix Frequencies x 100 A R N D 2 1 1 1 8 19 1 1 3 2 7 2 8 13 13 14 4 1 7 7 5 2 8 23 2 1 7 13 3 7 7 8 3 2 7 19 C Q E 1 1 1 7 2 2 1 17 13 1 2 13 7 1 2 1 6 3 1 1 2 7 1 6 2 G 1 2 2 2 8 8 H I L K M F P S T W Y V 1 4 16 1 6 15 7 1 2 7 18 13 1 3 15 13 6 2 1 2 2 7 1 8 14 3 1 1 7 7 2 0 1 8 1 2 3 3 1 7 1 3 7 0 1 7 1 13 15 2 6 6 1 7 2 7 7 4 1 3 3 2 1 13 8 0 1 18 1 8 14 2 6 1 20 7 2 7 1 3 1 2 8 2 1 1 13 7 2 7 1 9 9 2 1 1 1 7 2 0 1 18

More on Logos l Information content D = S pi log 2 (pi/qi) l Shannon, qi = 1/N = 0. 05 D = S pi log 2 (pi) - S pi log 2 (1/N) = log 2 N - S pi log 2 (pi) l Kullback-Leibler, qi = background frequency – V/L/A more frequent than for instance C/H/W
![Mutual information P 1 Ii j Saai Saaj Paai aaj logPaai aajPaaiPaaj Mutual information P 1 I(i, j) = Saai Saaj P(aai, aaj) * log[P(aai, aaj)/P(aai)*P(aaj)]](https://slidetodoc.com/presentation_image_h2/b073db05fba5248a1f1455d0d80fe454/image-12.jpg)
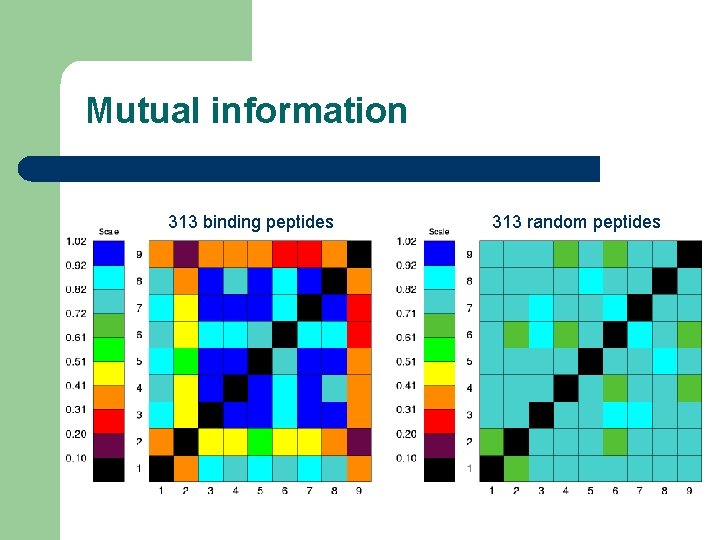
Mutual information P 1 I(i, j) = Saai Saaj P(aai, aaj) * log[P(aai, aaj)/P(aai)*P(aaj)] P(G 1) = 2/9 = 0. 22, . . P(V 6) = 4/9 = 0. 44, . . P(G 1, V 6) = 2/9 = 0. 22, P(G 1)*P(V 6) = 8/81 = 0. 10 log(0. 22/0. 10) > 0 P 6 ALWGFFPVA ILKEPVHGV ILGFVFTLT LLFGYPVYV GLSPTVWLS YMNGTMSQV GILGFVFTL WLSLLVPFV FLPSDFFPS

Mutual information 313 binding peptides 313 random peptides

Weight matrices l l Estimate amino acid frequencies from alignment inc. sequence weighting and pseudo counts Now a weight matrix is given as Wij = log(pij/qj) l l l Here i is a position in the motif, and j an amino acid. qj is the background frequency for amino acid j. W is a L x 20 matrix, L is motif length Score sequences to weight matrix by looking up and adding L values from matrix

Example from real life l l l 10 peptides from MHCpep database Bind to the MHC complex Relevant for immune system recognition Estimate sequence motif and weight matrix Evaluate on 528 peptides ALAKAAAAM ALAKAAAAN ALAKAAAAR ALAKAAAAT ALAKAAAAV GMNERPILT GILGFVFTM TLNAWVKVV KLNEPVLLL AVVPFIVSV

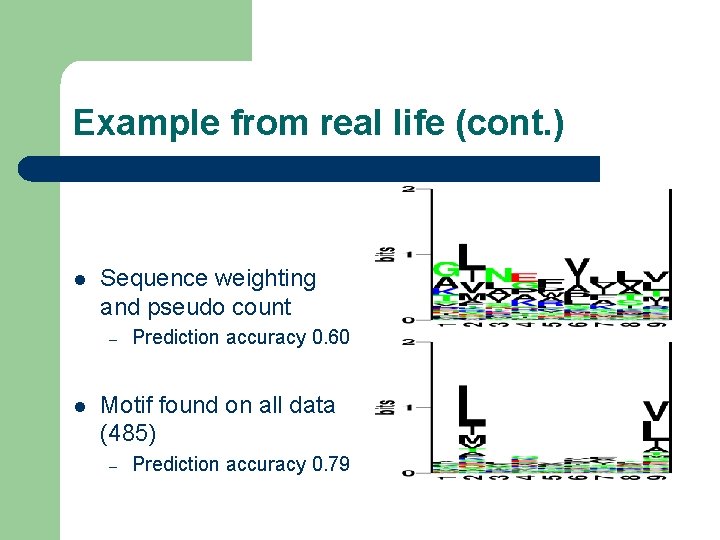
Example from real life (cont. ) l Raw sequence counting – – – l No sequence weighting No pseudo count Prediction accuracy 0. 45 Sequence weighting – – No pseudo count Prediction accuracy 0. 5

Example from real life (cont. ) l Sequence weighting and pseudo count – l Prediction accuracy 0. 60 Motif found on all data (485) – Prediction accuracy 0. 79

Hidden Markov Models l l l Weight matrices do not deal with insertions and deletions In alignments, this is done in an ad-hoc manner by optimization of the two gap penalties for first gap and gap extension HMM is a natural frame work where insertions/deletions are dealt with explicitly

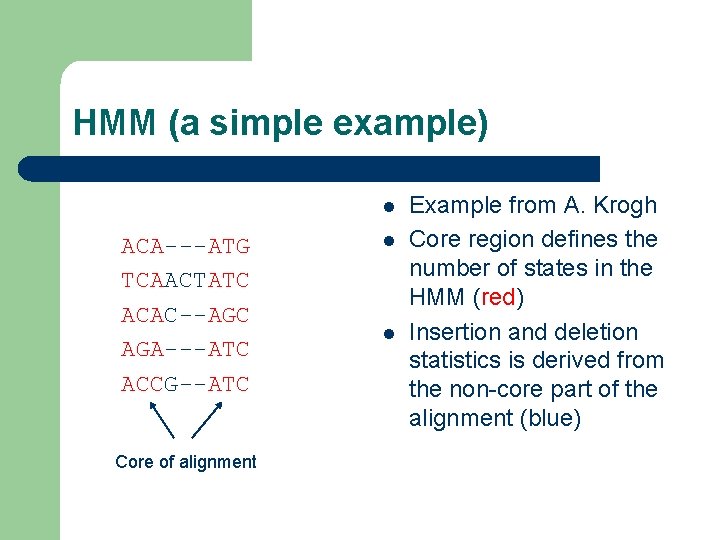
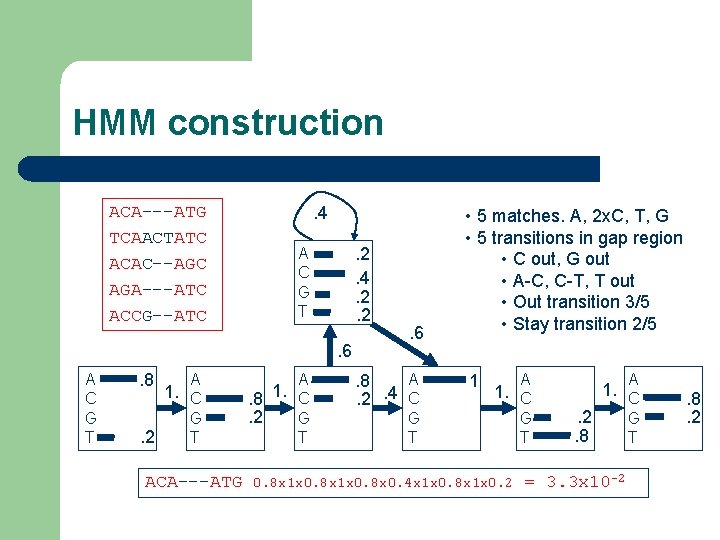
HMM (a simple example) l ACA---ATG TCAACTATC ACAC--AGC AGA---ATC ACCG--ATC Core of alignment l l Example from A. Krogh Core region defines the number of states in the HMM (red) Insertion and deletion statistics is derived from the non-core part of the alignment (blue)

HMM construction ACA---ATG TCAACTATC ACAC--AGC AGA---ATC ACCG--ATC . 4. 2. 2 A C G T . 6 A C G T . 8. 2 A 1. C G T ACA---ATG A . 8 1. C. 2 G T . 6 A. 8. 4 C. 2 • 5 matches. A, 2 x. C, T, G • 5 transitions in gap region • C out, G out • A-C, C-T, T out • Out transition 3/5 • Stay transition 2/5 1 A 1. C G T 0. 8 x 1 x 0. 8 x 0. 4 x 1 x 0. 8 x 1 x 0. 2 G T A . 2. 8 1. C = 3. 3 x 10 -2 G T . 8. 2

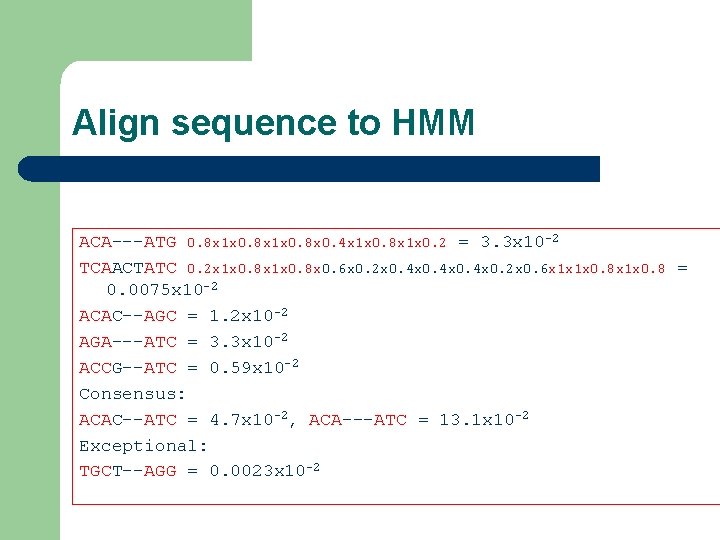
Align sequence to HMM ACA---ATG 0. 8 x 1 x 0. 8 x 0. 4 x 1 x 0. 8 x 1 x 0. 2 = 3. 3 x 10 -2 TCAACTATC 0. 2 x 1 x 0. 8 x 0. 6 x 0. 2 x 0. 4 x 0. 2 x 0. 6 x 1 x 1 x 0. 8 = 0. 0075 x 10 -2 ACAC--AGC = 1. 2 x 10 -2 AGA---ATC = 3. 3 x 10 -2 ACCG--ATC = 0. 59 x 10 -2 Consensus: ACAC--ATC = 4. 7 x 10 -2, ACA---ATC = 13. 1 x 10 -2 Exceptional: TGCT--AGG = 0. 0023 x 10 -2

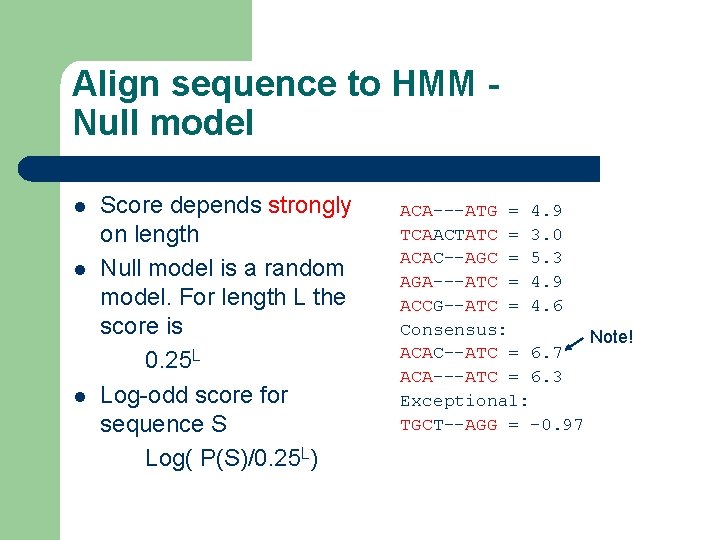
Align sequence to HMM Null model l Score depends strongly on length Null model is a random model. For length L the score is 0. 25 L Log-odd score for sequence S Log( P(S)/0. 25 L) ACA---ATG = 4. 9 TCAACTATC = 3. 0 ACAC--AGC = 5. 3 AGA---ATC = 4. 9 ACCG--ATC = 4. 6 Consensus: Note! ACAC--ATC = 6. 7 ACA---ATC = 6. 3 Exceptional: TGCT--AGG = -0. 97

HMM’s and weight matrices l l Note. In the case of un-gapped alignments HMM’s become simple weight matrices It still might be useful to use a HMM tool package to estimate a weight matrix – – Sequence weighting Pseudo counts

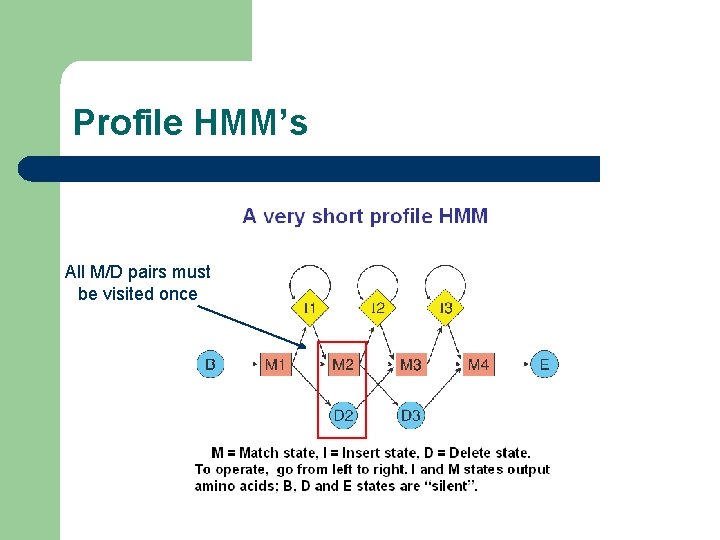
Profile HMM’s n io ert Ins EM 55_HUMAN CSKP_HUMAN KAPB_MOUSE NRC 2_NEUCR WWQGRVEGSSKESAGLIPSPELQEWRVASMAQSAP--SEAPSCSPFGKKKK-YKDKYLAK WWQGKLENSKNGTAGLIPSPELQEWRVACIAMEKTKQEQQASCTWFGKKKKQYKDKYLAK -----PENLLIDHQGYIQVTDFGFAKRVKG-----------------PENILLHQSGHIMLSDFDLSKQSDPGGKPTMIIGKNGTSTSSLPTIDTKSCIANF EM 55_HUMAN CSKP_HUMAN KAPB_MOUSE NRC 2_NEUCR HSSIFDQLDVVSYEEVVRLPAFKRKTLVLIGASGVGRSHIKNALLSQNPEKFVYPVPYTT HNAVFDQLDLVTYEEVVKLPAFKRKTLVLLGAHGVGRRHIKNTLITKHPDRFAYPIPHTT RTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG ion t RTNSFVGTEEYIAPEVIKGSGHTSAVDWWTLGILIYEMLYGTTPFKGKNRNATFANILRE e l EM 55_HUMAN CSKP_HUMAN KAPB_MOUSE NRC 2_NEUCR RPPRKSEEDGKEYHFISTEEMTRNISANEFLEFGSYQGNMFGTKFETVHQIHKQNKIAIL RPPKKDEENGKNYYFVSHDQMMQDISNNEYLEYGSHEDAMYGTKLETIRKIHEQGLIAIL KVRFPSHF-----SSDLKDLLRNLLQVDLTKRFGNLKNGVSDIKTHKWFATTDWIAIYQR DIPFPDHAGAPQISNLCKSLIRKLLIKDENRRLG-ARAGASDIKTHPFFRTTQWALI--R EM 55_HUMAN CSKP_HUMAN KAPB_MOUSE NRC 2_NEUCR NNGVDETLKKLQEAFDQACSSPQWVPVSWVY NNEIDETIRHLEEAVELVCTAPQWVPVSWVY EKCGKEFCEF----------ENAVDPFEEFNSVTLHHDGDEEYHSDAYEKR De

Profile HMM’s All M/D pairs must be visited once

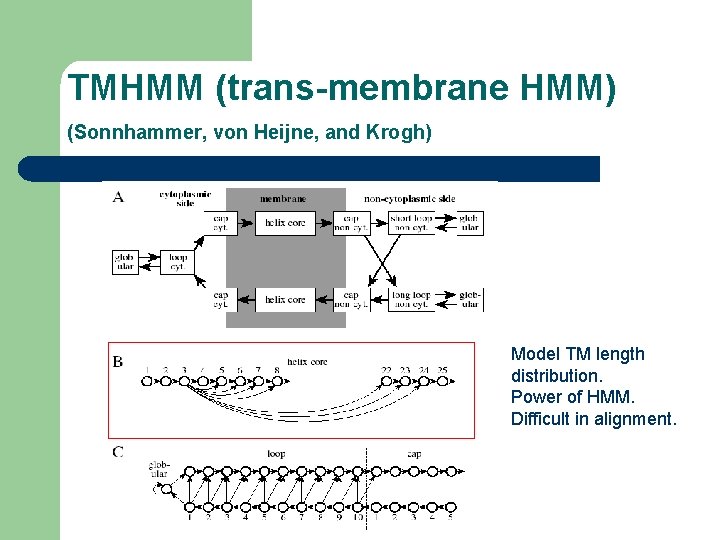
TMHMM (trans-membrane HMM) (Sonnhammer, von Heijne, and Krogh) Model TM length distribution. Power of HMM. Difficult in alignment.

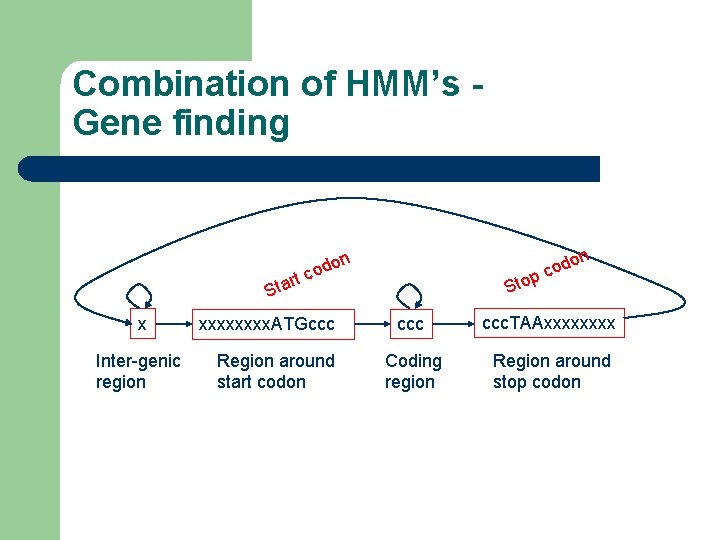
Combination of HMM’s Gene finding rt Sta x Inter-genic region on d o c xxxx. ATGccc Region around start codon p Sto o cod n ccc. TAAxxxx Coding region Region around stop codon

HMM packages l l HMMER (http: //hmmer. wustl. edu/) – S. R. Eddy, Wash. U St. Louis. Freely available. SAM (http: //www. cse. ucsc. edu/research/compbio/sam. html) – R. Hughey, K. Karplus, A. Krogh, D. Haussler and others, UC Santa Cruz. Freely available to academia, nominal license fee for commercial users. META-MEME (http: //metameme. sdsc. edu/) – William Noble Grundy, UC San Diego. Freely available. Combines features of PSSM search and profile HMM search. NET-ID, HMMpro (http: //www. netid. com/html/hmmpro. html) – – Freely available to academia, nominal license fee for commercial users. Allows HMM architecture construction.

Simple Hmmer command hmmbuild --gapmax 0. 0 --fast A 2. hmmer A 2. fsa >HLA-A. 0201 16 Example_for_Ligand SLLPAIVEL >HLA-A. 0201 16 Example_for_Ligand YLLPAIVHI >HLA-A. 0201 16 Example_for_Ligand TLWVDPYEV >HLA-A. 0201 16 Example_for_Ligand SXPSGGXGV >HLA-A. 0201 16 Example_for_Ligand GLVPFLVSV hmmbuild - build a hidden Markov model from an alignment HMMER 2. 2 g (August 2001) ------------------Alignment file: A 2. fsa File format: a 2 m Search algorithm configuration: Multiple domain (hmmls) Model construction strategy: Fast/ad hoc (gapmax 0. 0) Null model used: (default) Sequence weighting method: G/S/C tree weights ----------------Alignment: #1 Number of sequences: 232 Number of columns: 9 Determining effective sequence number. . . done. [192] Weighting sequences heuristically. . . done. Constructing model architecture. . . done. Converting counts to probabilities. . . done. Setting model name, etc. . done. [A 2. fasta] Constructed a profile HMM (length 9) Average score: -6. 42 bits Minimum score: -15. 47 bits Maximum score: -0. 84 bits Std. deviation: 2. 72 bits