Ph D defence KATHOLIEKE UNIVERSITEI T LEUVEN Realization

![Finding motifs in DNA sequences Find motifs in muscle specific regulatory regions [Zhou, Wong] Finding motifs in DNA sequences Find motifs in muscle specific regulatory regions [Zhou, Wong]](https://slidetodoc.com/presentation_image_h2/8247431f5aa23f3bd5d36b41cb0b8e5b/image-40.jpg)


- Slides: 44

Ph. D. defence KATHOLIEKE UNIVERSITEI T LEUVEN Realization, identification and filtering for hidden Markov models using matrix factorization techniques Bart Vanluyten Promotoren: Prof. dr. ir. Bart De Moor, promotor Prof. dr. ir. Jan Willems, copromotor Juryleden: Maandag 5 mei 2008 Prof. dr. ir. H. Van Brussel, voorzitter Prof. dr. A. Bultheel Prof. dr. V. Blondel (UCL, Louvain-la-Neuve) Prof. dr. P. Spreij (UVA, Amsterdam) Prof. dr. ir. L. Finesso (ISIB-CNR, Padova) Prof. dr. ir. K. Meerbergen

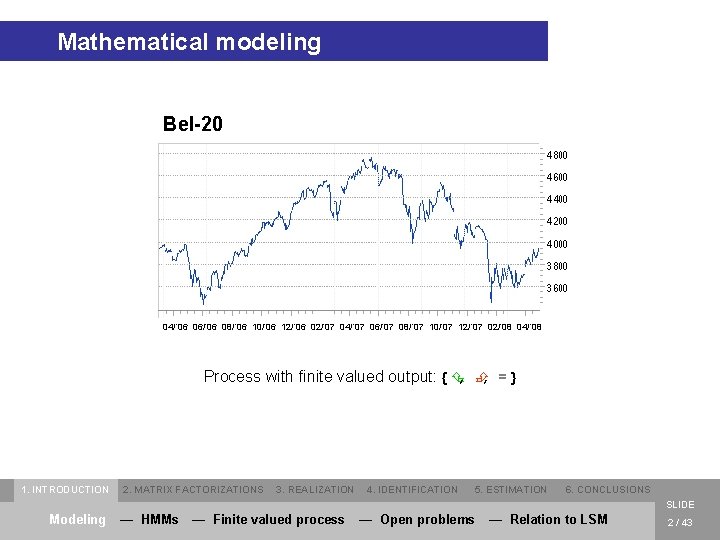
Mathematical modeling Bel-20 04/’ 06 06/’ 06 08/’ 06 10/’ 06 12/’ 06 02/’ 07 04/’ 07 06/’ 07 08/’ 07 10/’ 07 12/’ 07 02/’ 08 04/’ 08 Process with finite valued output: { Ç, È, = } 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Modeling — HMMs — Finite valued process — Open problems — Relation to LSM 2 / 43

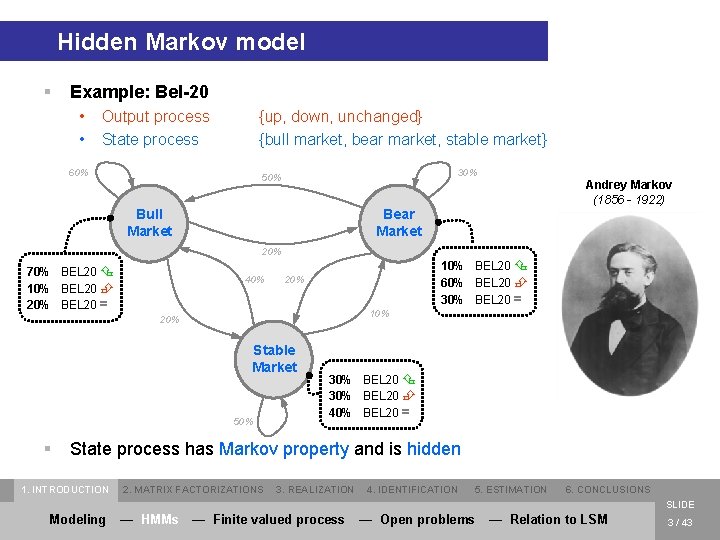
Hidden Markov model § Example: Bel-20 • • Output process State process {up, down, unchanged} {bull market, bear market, stable market} 60% 30% 50% Andrey Markov (1856 - 1922) Bull Market Bear Market 20% 70% BEL 20 Ç 10% BEL 20 È 20% BEL 20 = 40% 10% BEL 20 Ç 60% BEL 20 È 30% BEL 20 = 20% 10% 20% Stable Market 50% § 30% BEL 20 Ç 30% BEL 20 È 40% BEL 20 = State process has Markov property and is hidden 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Modeling — HMMs — Finite valued process — Open problems — Relation to LSM 3 / 43

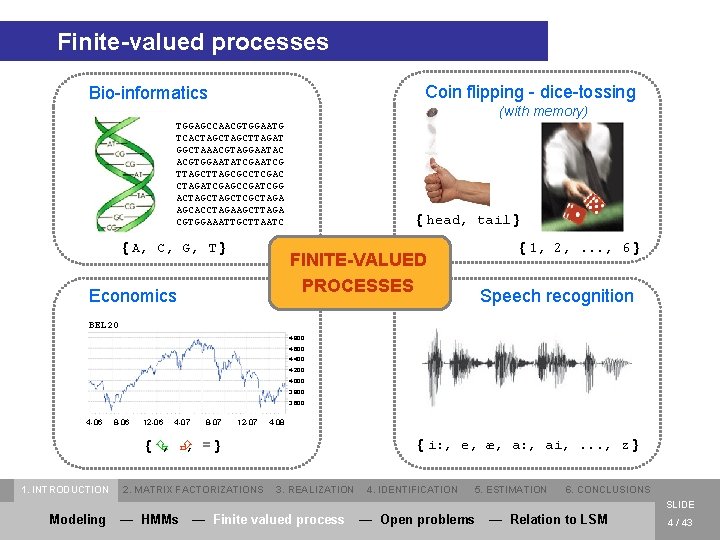
Finite-valued processes Coin flipping - dice-tossing Bio-informatics (with memory) TGGAGCCAACGTGGAATG TCACTAGCTTAGAT GGCTAAACGTAGGAATAC ACGTGGAATATCGAATCG TTAGCGCCTCGAC CTAGATCGAGCCGATCGG ACTAGCTCGCTAGA AGCACCTAGAAGCTTAGA CGTGGAAATTGCTTAATC { A, C, G, T } { head, tail } { 1, 2, . . . , 6 } FINITE-VALUED PROCESSES Economics Speech recognition BEL 20 4. 800 4. 600 4. 400 4. 200 4. 000 3. 800 3. 600 4 -06 8 -06 12 -06 4 -07 8 -07 12 -07 4 -08 { i: , e, æ, a: , ai, . . . , z } {Ç , È , =} 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Modeling — HMMs — Finite valued process — Open problems — Relation to LSM 4 / 43

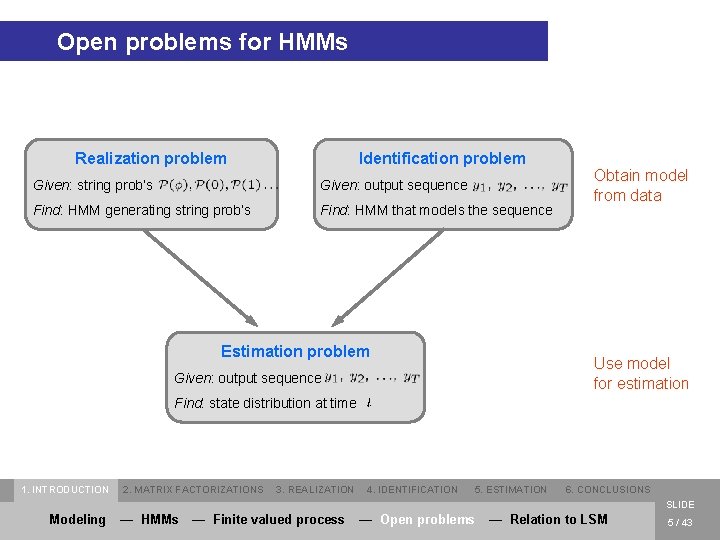
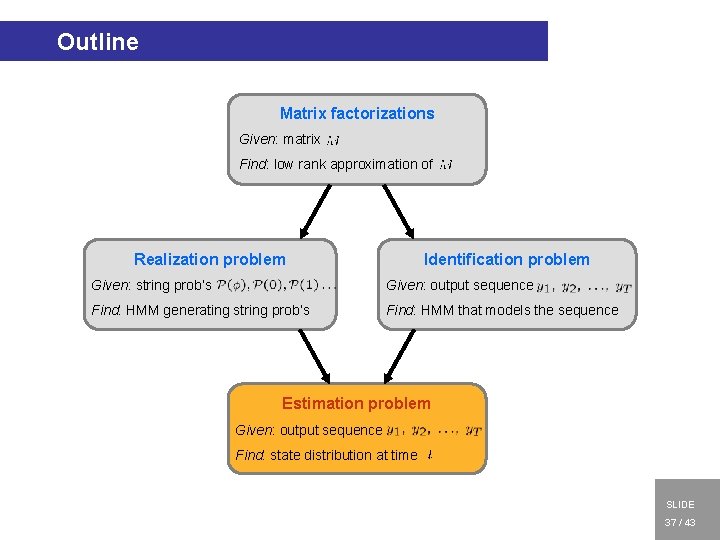
Open problems for HMMs Realization problem Identification problem Given: string prob’s Given: output sequence Find: HMM generating string prob’s Find: HMM that models the sequence Estimation problem Obtain model from data Use model for estimation Given: output sequence Find: state distribution at time 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Modeling — HMMs — Finite valued process — Open problems — Relation to LSM 5 / 43

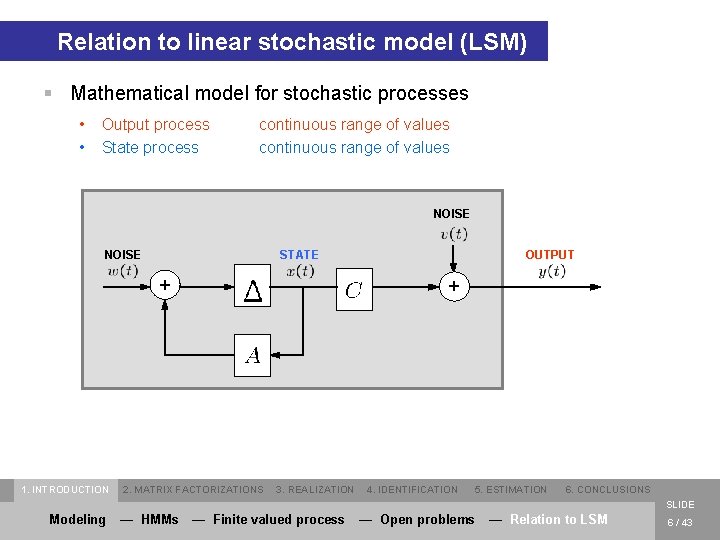
Relation to linear stochastic model (LSM) § Mathematical model for stochastic processes • • Output process State process continuous range of values NOISE STATE + 1. INTRODUCTION OUTPUT + 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Modeling — HMMs — Finite valued process — Open problems — Relation to LSM 6 / 43

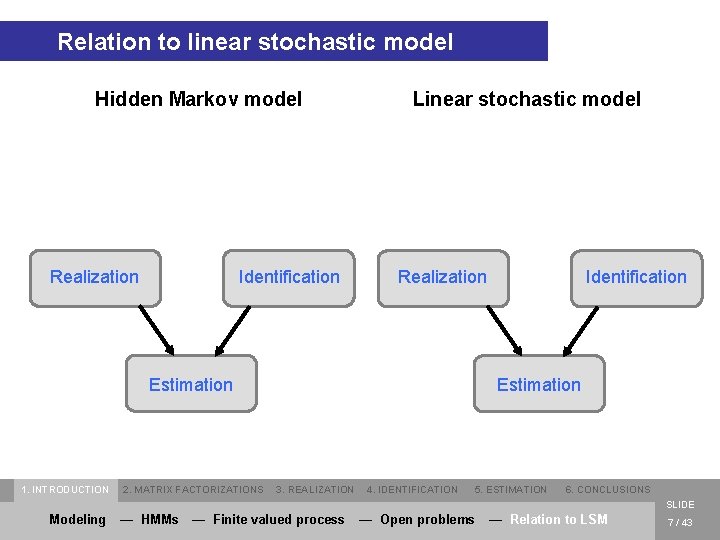
Relation to linear stochastic model Hidden Markov model Realization Identification Linear stochastic model Realization Estimation 1. INTRODUCTION 2. MATRIX FACTORIZATIONS Identification Estimation 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Modeling — HMMs — Finite valued process — Open problems — Relation to LSM 7 / 43

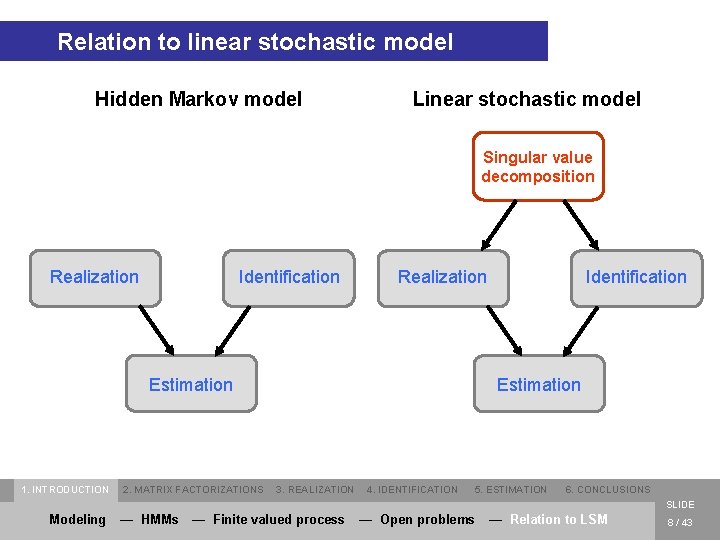
Relation to linear stochastic model Hidden Markov model Linear stochastic model Singular value decomposition Realization Identification Realization Estimation 1. INTRODUCTION 2. MATRIX FACTORIZATIONS Identification Estimation 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Modeling — HMMs — Finite valued process — Open problems — Relation to LSM 8 / 43

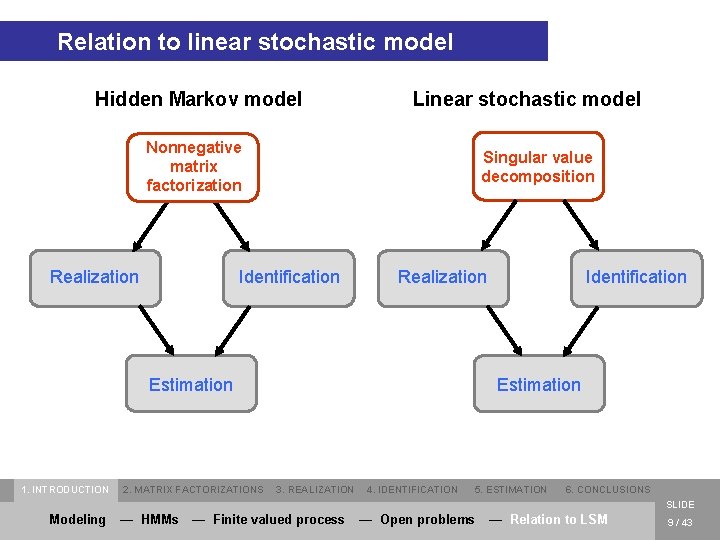
Relation to linear stochastic model Hidden Markov model Linear stochastic model Nonnegative matrix factorization Realization Singular value decomposition Identification Realization Estimation 1. INTRODUCTION 2. MATRIX FACTORIZATIONS Identification Estimation 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Modeling — HMMs — Finite valued process — Open problems — Relation to LSM 9 / 43

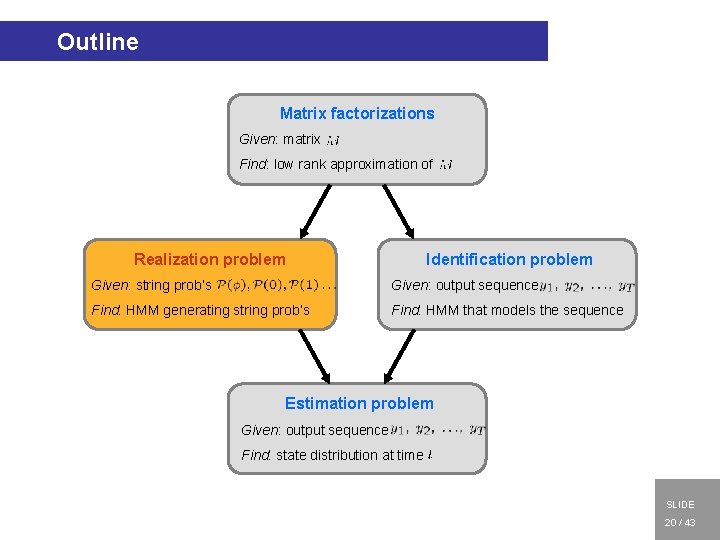
Outline Matrix factorizations 2 nd objective Given: matrix Find: low rank approximation of Realization problem Identification problem Given: string prob’s Given: output sequence Find: HMM generating string prob’s Find: HMM that models the sequence 1 st objective Estimation problem Given: output sequence Find: state distribution at time 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE 10 / 43

Outline Matrix factorizations Given: matrix Find: low rank approximation of Realization problem Identification problem Given: string prob’s Given: output sequence Find: HMM generating string prob’s Find: HMM that models the sequence Estimation problem Given: output sequence Find: state distribution at time 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE 11 / 43

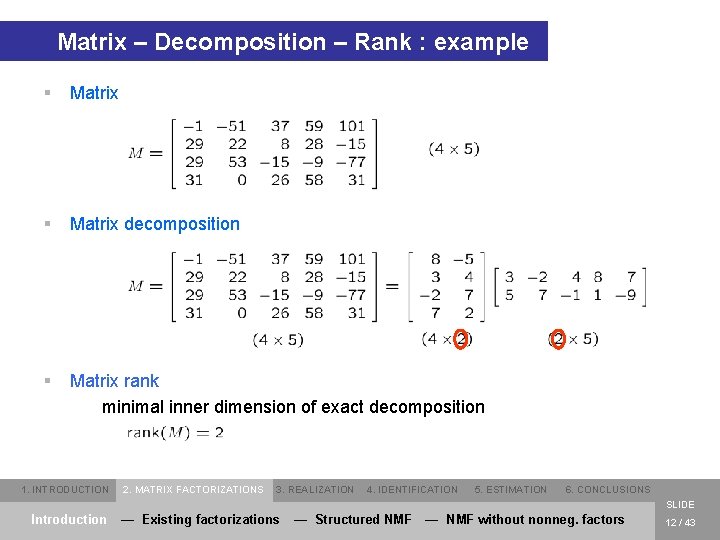
Matrix – Decomposition – Rank : example § Matrix decomposition § Matrix rank minimal inner dimension of exact decomposition 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Existing factorizations — Structured NMF — NMF without nonneg. factors 12 / 43

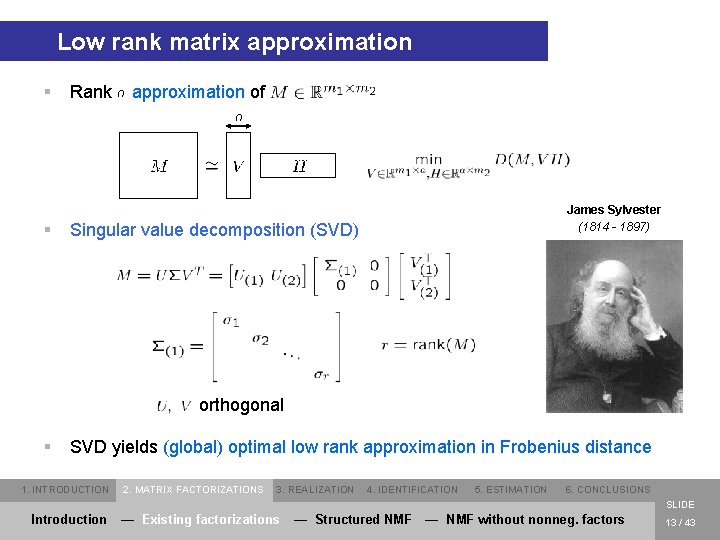
Low rank matrix approximation § § Rank approximation of James Sylvester (1814 - 1897) Singular value decomposition (SVD) orthogonal § SVD yields (global) optimal low rank approximation in Frobenius distance 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Existing factorizations — Structured NMF — NMF without nonneg. factors 13 / 43

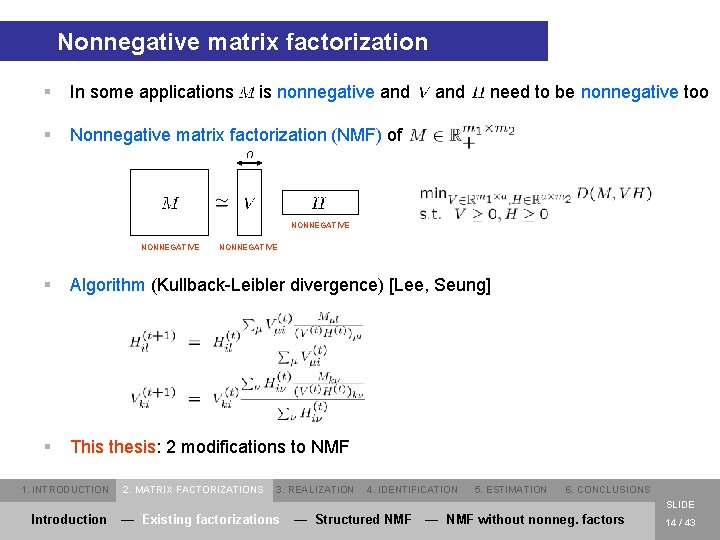
Nonnegative matrix factorization § In some applications § Nonnegative matrix factorization (NMF) of is nonnegative and need to be nonnegative too NONNEGATIVE § Algorithm (Kullback-Leibler divergence) [Lee, Seung] § This thesis: 2 modifications to NMF 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Existing factorizations — Structured NMF — NMF without nonneg. factors 14 / 43

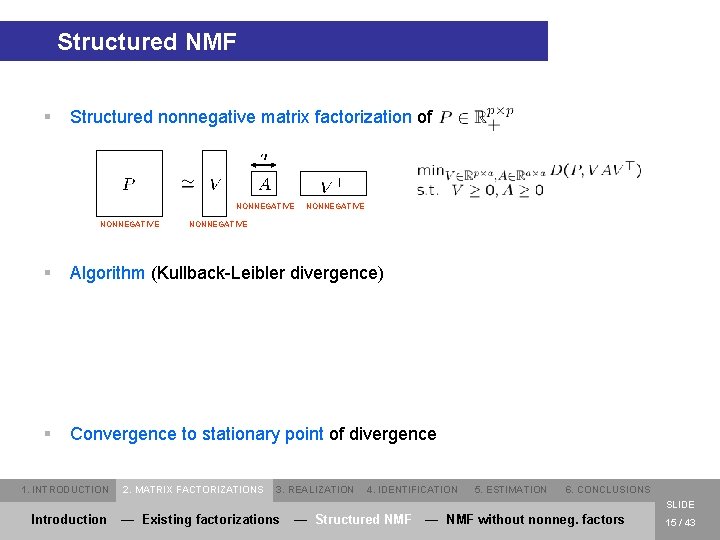
Structured NMF § Structured nonnegative matrix factorization of NONNEGATIVE § Algorithm (Kullback-Leibler divergence) § Convergence to stationary point of divergence 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Existing factorizations — Structured NMF — NMF without nonneg. factors 15 / 43

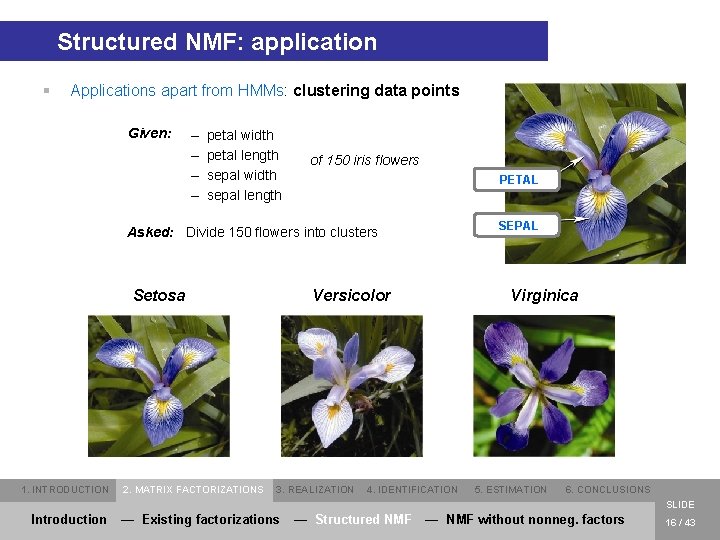
Structured NMF: application § Applications apart from HMMs: clustering data points Given: – – petal width petal length sepal width sepal length of 150 iris flowers PETAL SEPAL Asked: Divide 150 flowers into clusters Setosa 1. INTRODUCTION 2. MATRIX FACTORIZATIONS Versicolor 3. REALIZATION Virginica 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Existing factorizations — Structured NMF — NMF without nonneg. factors 16 / 43

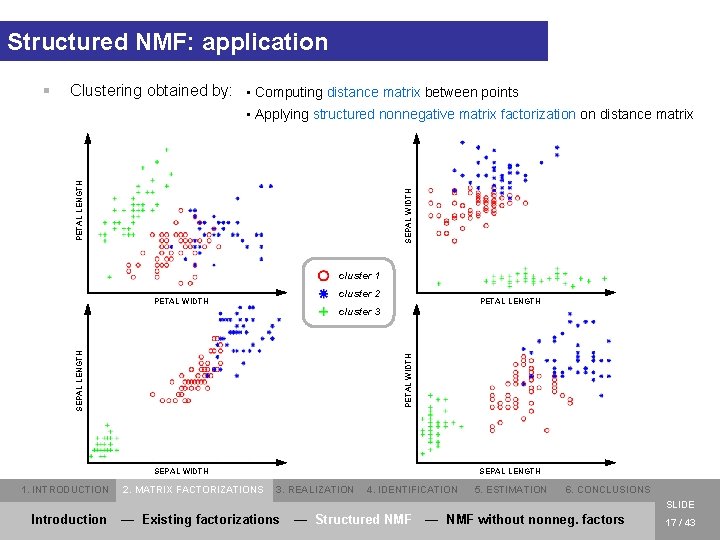
Structured NMF: application § Clustering obtained by: • Computing distance matrix between points SEPAL WIDTH PETAL LENGTH • Applying structured nonnegative matrix factorization on distance matrix cluster 1 cluster 2 PETAL WIDTH PETAL LENGTH PETAL WIDTH SEPAL LENGTH cluster 3 SEPAL WIDTH 1. INTRODUCTION 2. MATRIX FACTORIZATIONS SEPAL LENGTH 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Existing factorizations — Structured NMF — NMF without nonneg. factors 17 / 43

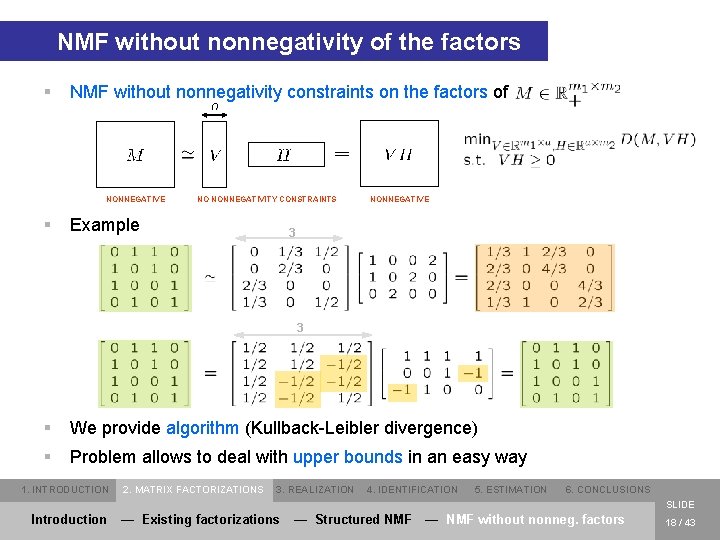
NMF without nonnegativity of the factors § NMF without nonnegativity constraints on the factors of NONNEGATIVE § NO NONNEGATIVITY CONSTRAINTS Example NONNEGATIVE 3 3 § We provide algorithm (Kullback-Leibler divergence) § Problem allows to deal with upper bounds in an easy way 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Existing factorizations — Structured NMF — NMF without nonneg. factors 18 / 43

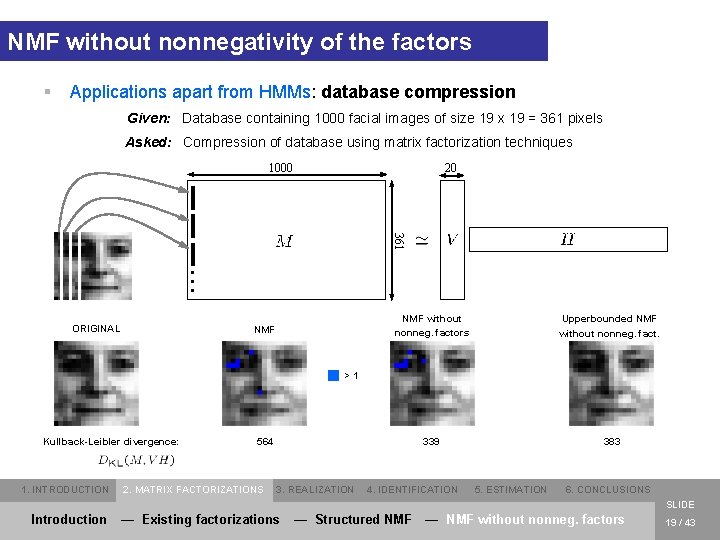
NMF without nonnegativity of the factors § Applications apart from HMMs: database compression Given: Database containing 1000 facial images of size 19 x 19 = 361 pixels Asked: Compression of database using matrix factorization techniques 20 1000 . . . 361 ORIGINAL NMF without nonneg. factors Upperbounded NMF without nonneg. fact. 339 383 >1 Kullback-Leibler divergence: 1. INTRODUCTION 564 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Existing factorizations — Structured NMF — NMF without nonneg. factors 19 / 43

Outline Matrix factorizations Given: matrix Find: low rank approximation of Realization problem Identification problem Given: string prob’s Given: output sequence Find: HMM generating string prob’s Find: HMM that models the sequence Estimation problem Given: output sequence Find: state distribution at time 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE 20 / 43

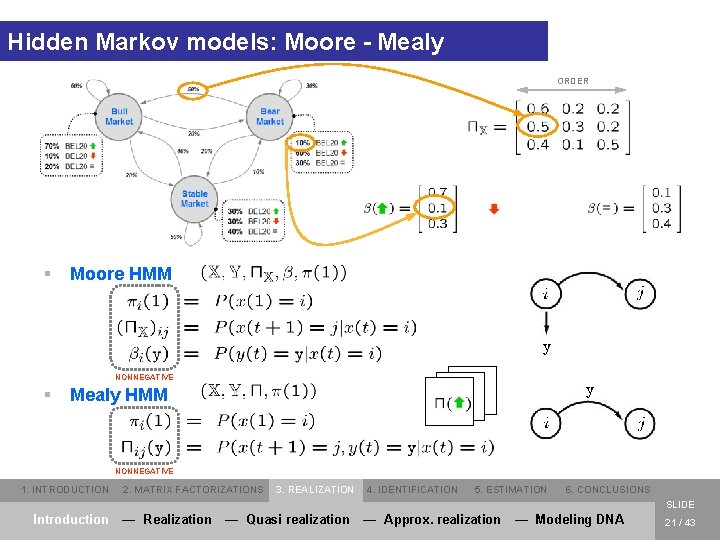
Hidden Markov models: Moore - Mealy ORDER = § Moore HMM NONNEGATIVE § Mealy HMM NONNEGATIVE 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Realization — Quasi realization — Approx. realization — Modeling DNA 21 / 43

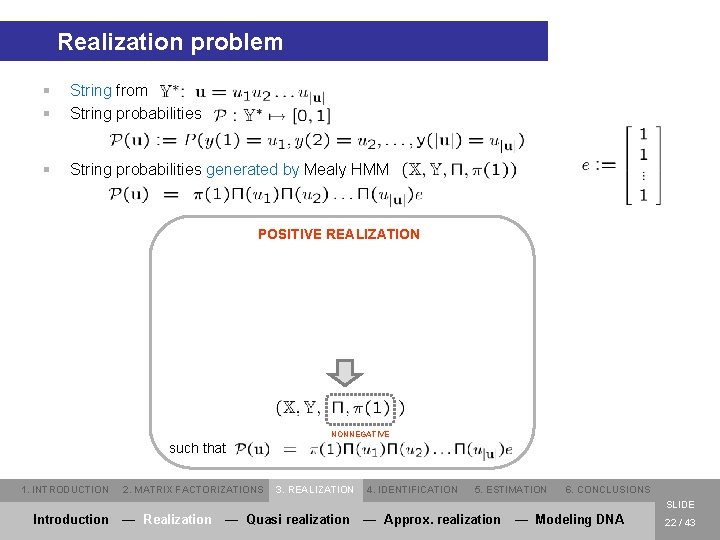
Realization problem § § String from String probabilities § String probabilities generated by Mealy HMM POSITIVE REALIZATION NONNEGATIVE such that 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Realization — Quasi realization — Approx. realization — Modeling DNA 22 / 43

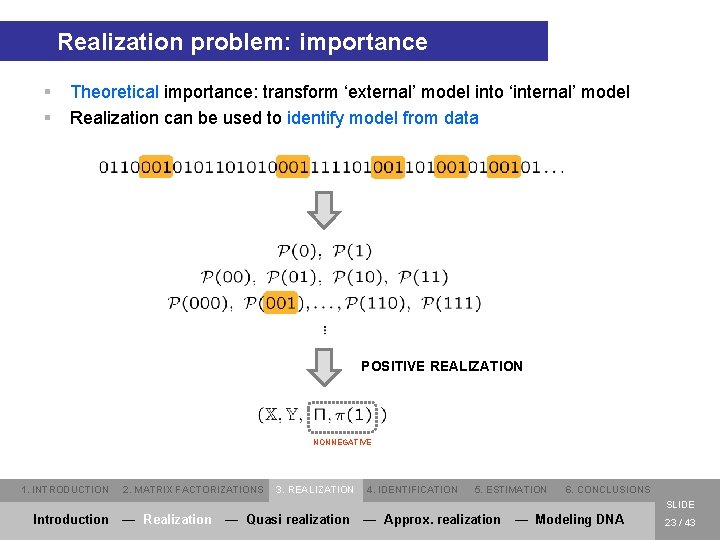
Realization problem: importance § § Theoretical importance: transform ‘external’ model into ‘internal’ model Realization can be used to identify model from data POSITIVE REALIZATION NONNEGATIVE 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Realization — Quasi realization — Approx. realization — Modeling DNA 23 / 43

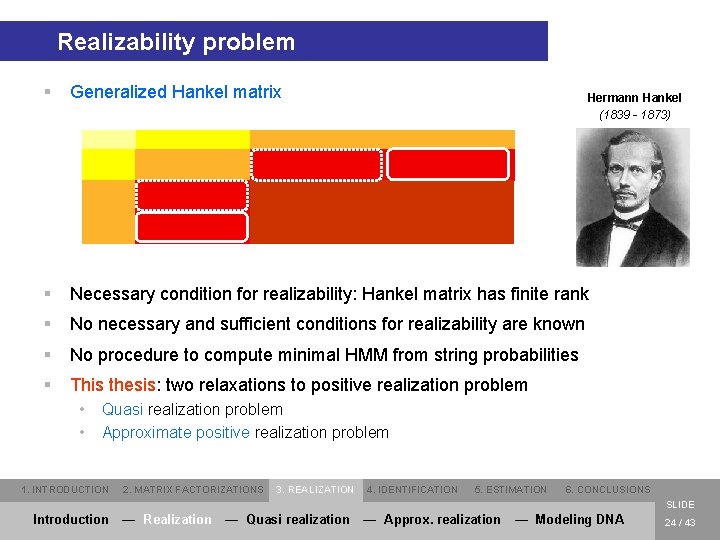
Realizability problem § Generalized Hankel matrix § Necessary condition for realizability: Hankel matrix has finite rank § No necessary and sufficient conditions for realizability are known § No procedure to compute minimal HMM from string probabilities § This thesis: two relaxations to positive realization problem • • Hermann Hankel (1839 - 1873) Quasi realization problem Approximate positive realization problem 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Realization — Quasi realization — Approx. realization — Modeling DNA 24 / 43

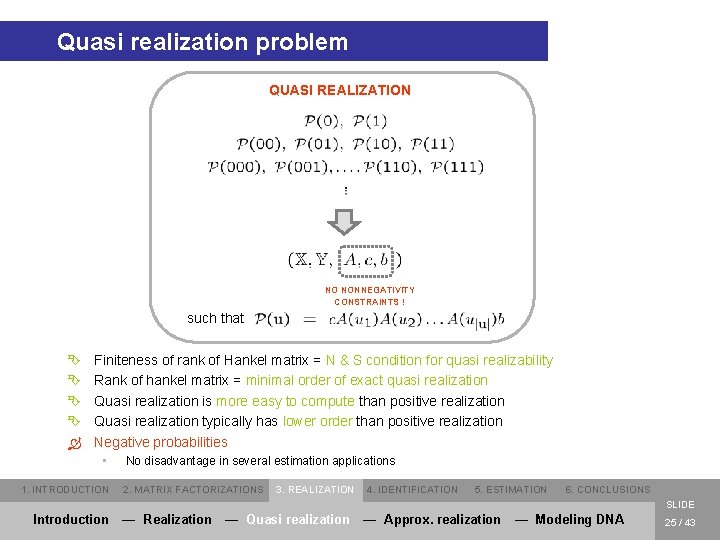
Quasi realization problem QUASI REALIZATION NO NONNEGATIVITY CONSTRAINTS ! such that Ê Ê Ò Finiteness of rank of Hankel matrix = N & S condition for quasi realizability Rank of hankel matrix = minimal order of exact quasi realization Quasi realization is more easy to compute than positive realization Quasi realization typically has lower order than positive realization Negative probabilities • 1. INTRODUCTION No disadvantage in several estimation applications 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Realization — Quasi realization — Approx. realization — Modeling DNA 25 / 43

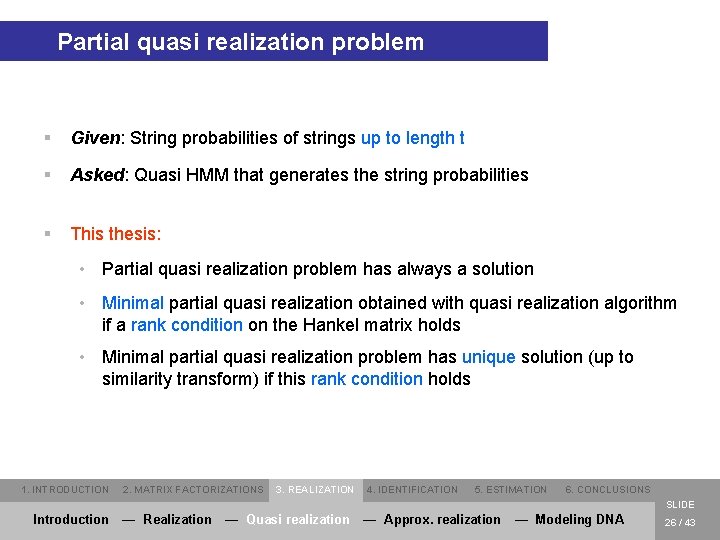
Partial quasi realization problem § Given: String probabilities of strings up to length t § Asked: Quasi HMM that generates the string probabilities § This thesis: • Partial quasi realization problem has always a solution • Minimal partial quasi realization obtained with quasi realization algorithm if a rank condition on the Hankel matrix holds • Minimal partial quasi realization problem has unique solution (up to similarity transform) if this rank condition holds 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Realization — Quasi realization — Approx. realization — Modeling DNA 26 / 43

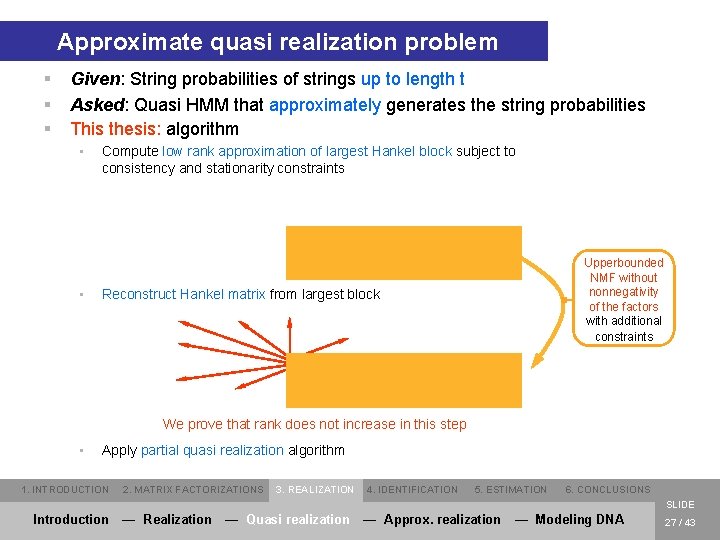
Approximate quasi realization problem § § § Given: String probabilities of strings up to length t Asked: Quasi HMM that approximately generates the string probabilities This thesis: algorithm • • Compute low rank approximation of largest Hankel block subject to consistency and stationarity constraints Upperbounded NMF without nonnegativity of the factors with additional constraints Reconstruct Hankel matrix from largest block We prove that rank does not increase in this step • Apply partial quasi realization algorithm 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Realization — Quasi realization — Approx. realization — Modeling DNA 27 / 43

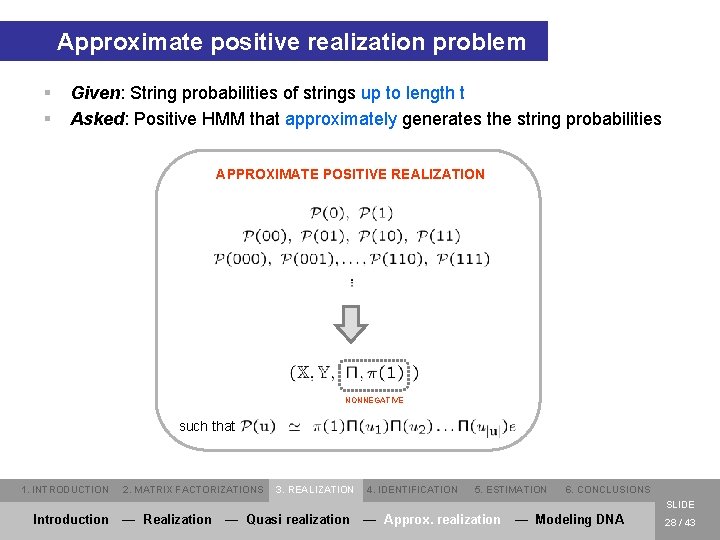
Approximate positive realization problem § § Given: String probabilities of strings up to length t Asked: Positive HMM that approximately generates the string probabilities APPROXIMATE POSITIVE REALIZATION NONNEGATIVE such that 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Realization — Quasi realization — Approx. realization — Modeling DNA 28 / 43

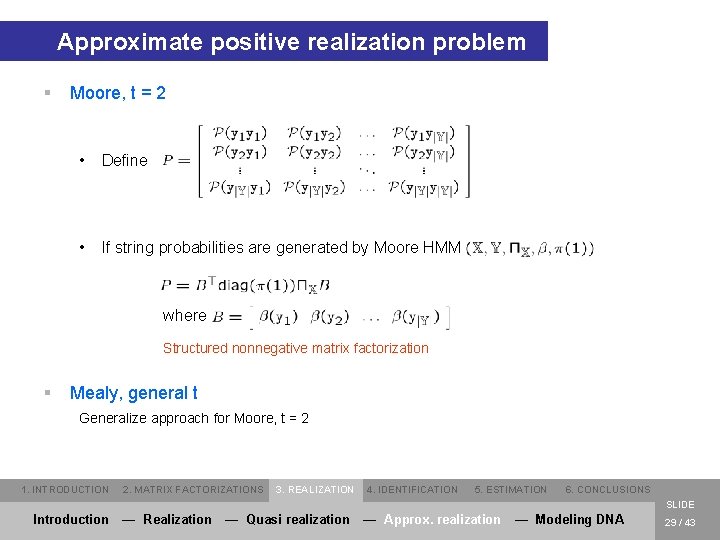
Approximate positive realization problem § Moore, t = 2 • Define • If string probabilities are generated by Moore HMM where Structured nonnegative matrix factorization § Mealy, general t Generalize approach for Moore, t = 2 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Realization — Quasi realization — Approx. realization — Modeling DNA 29 / 43

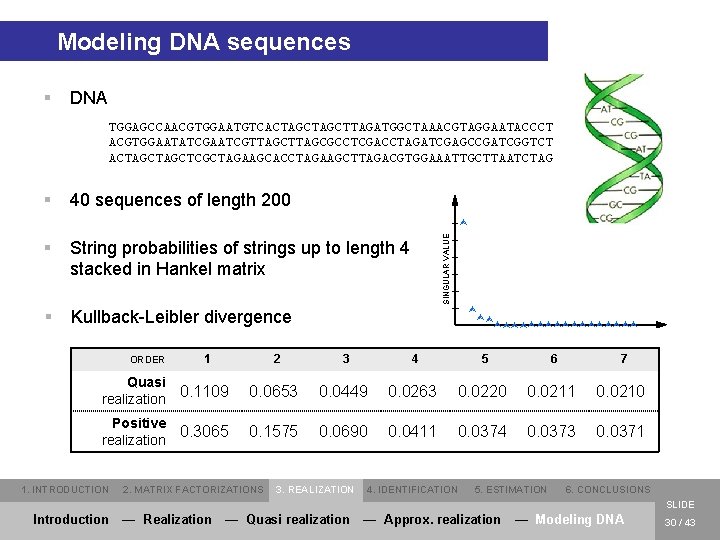
Modeling DNA sequences § DNA TGGAGCCAACGTGGAATGTCACTAGCTTAGATGGCTAAACGTAGGAATACCCT ACGTGGAATATCGAATCGTTAGCGCCTCGACCTAGATCGAGCCGATCGGTCT ACTAGCTCGCTAGAAGCACCTAGAAGCTTAGACGTGGAAATTGCTTAATCTAG § 40 sequences of length 200 § String probabilities of strings up to length 4 stacked in Hankel matrix § Kullback-Leibler divergence SINGULAR VALUE Ù Ù ÙÙ ÙÙÙÙÙÙÙÙÙ 1 2 3 4 5 6 7 Quasi realization 0. 1109 0. 0653 0. 0449 0. 0263 0. 0220 0. 0211 0. 0210 Positive realization 0. 3065 0. 1575 0. 0690 0. 0411 0. 0374 0. 0373 0. 0371 ORDER 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Realization — Quasi realization — Approx. realization — Modeling DNA 30 / 43

Outline Matrix factorizations Given: matrix Find: low rank approximation of Realization problem Identification problem Given: string prob’s Given: output sequence Find: HMM generating string prob’s Find: HMM that models the sequence Estimation problem Given: output sequence Find: state distribution at time 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE 31 / 43

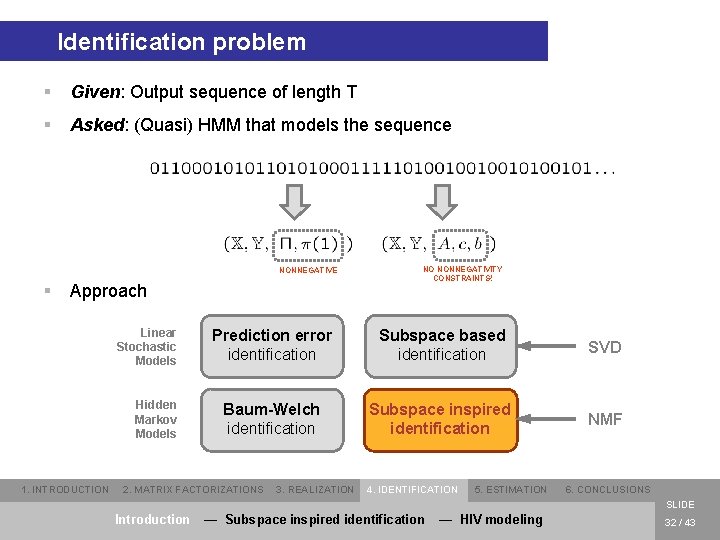
Identification problem § Given: Output sequence of length T § Asked: (Quasi) HMM that models the sequence NONNEGATIVE § Approach Linear Stochastic Models Hidden Markov Models 1. INTRODUCTION NO NONNEGATIVITY CONSTRAINTS! Prediction error identification Subspace based identification SVD Baum-Welch identification Subspace inspired identification NMF 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Subspace inspired identification — HIV modeling 32 / 43

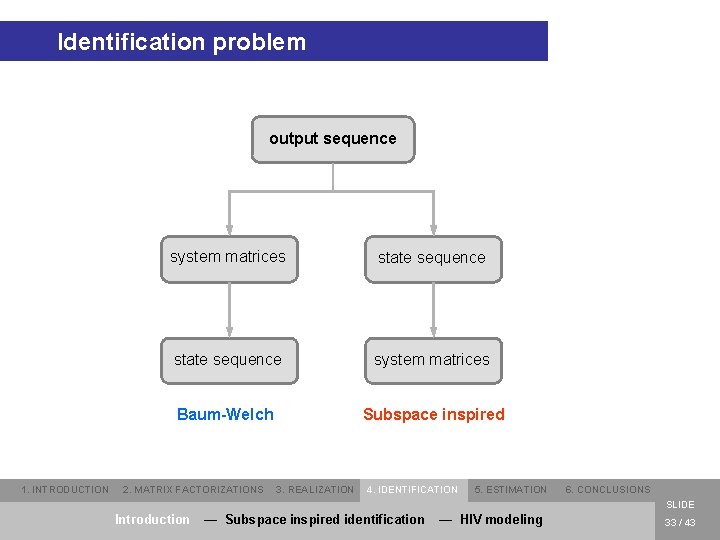
Identification problem output sequence 1. INTRODUCTION system matrices state sequence system matrices Baum-Welch Subspace inspired 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Subspace inspired identification — HIV modeling 33 / 43

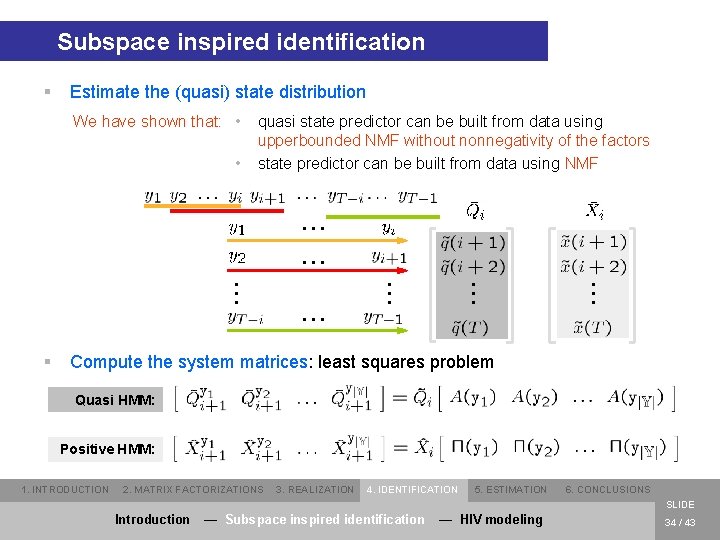
Subspace inspired identification § Estimate the (quasi) state distribution We have shown that: • • quasi state predictor can be built from data using upperbounded NMF without nonnegativity of the factors state predictor can be built from data using NMF . . . § . . . . Compute the system matrices: least squares problem Quasi HMM: Positive HMM: 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Subspace inspired identification — HIV modeling 34 / 43

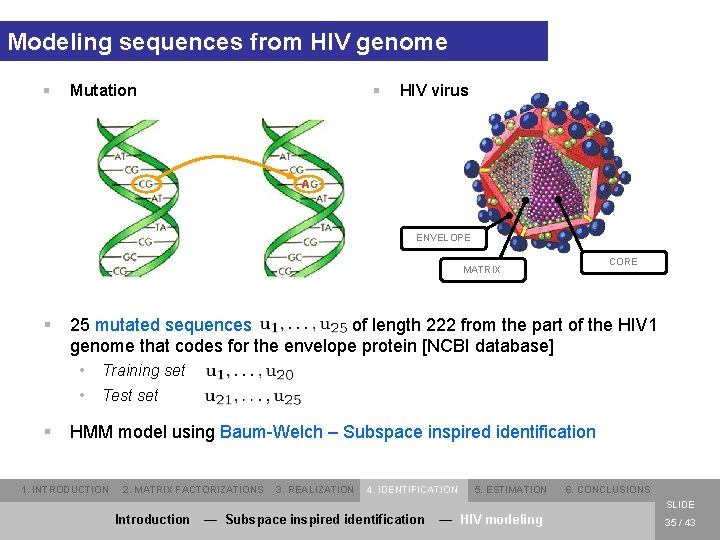
Modeling sequences from HIV genome § § Mutation HIV virus A ENVELOPE CORE MATRIX § § 25 mutated sequences of length 222 from the part of the HIV 1 genome that codes for the envelope protein [NCBI database] • Training set • Test set HMM model using Baum-Welch – Subspace inspired identification 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Subspace inspired identification — HIV modeling 35 / 43

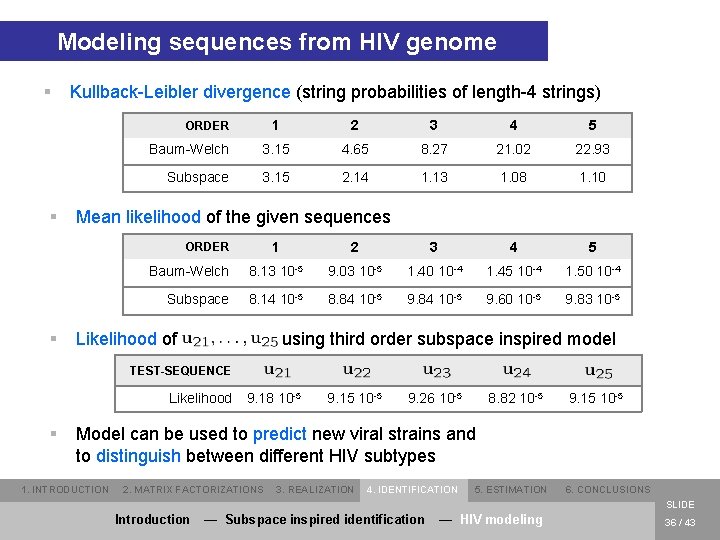
Modeling sequences from HIV genome § Kullback-Leibler divergence (string probabilities of length-4 strings) 1 2 3 4 5 Baum-Welch 3. 15 4. 65 8. 27 21. 02 22. 93 Subspace 3. 15 2. 14 1. 13 1. 08 1. 10 ORDER § Mean likelihood of the given sequences 1 2 3 4 5 Baum-Welch 8. 13 10 -5 9. 03 10 -5 1. 40 10 -4 1. 45 10 -4 1. 50 10 -4 Subspace 8. 14 10 -5 8. 84 10 -5 9. 60 10 -5 9. 83 10 -5 ORDER § Likelihood of using third order subspace inspired model TEST-SEQUENCE Likelihood § 9. 18 10 -5 9. 15 10 -5 9. 26 10 -5 8. 82 10 -5 9. 15 10 -5 Model can be used to predict new viral strains and to distinguish between different HIV subtypes 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Introduction — Subspace inspired identification — HIV modeling 36 / 43

Outline Matrix factorizations Given: matrix Find: low rank approximation of Realization problem Identification problem Given: string prob’s Given: output sequence Find: HMM generating string prob’s Find: HMM that models the sequence Estimation problem Given: output sequence Find: state distribution at time 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE 37 / 43

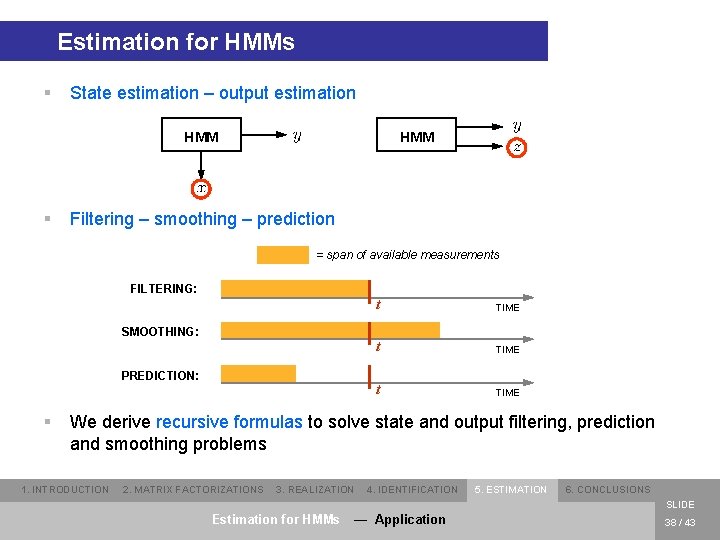
Estimation for HMMs § State estimation – output estimation HMM § HMM Filtering – smoothing – prediction = span of available measurements FILTERING: SMOOTHING: PREDICTION: § t TIME We derive recursive formulas to solve state and output filtering, prediction and smoothing problems 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Estimation for HMMs — Application 38 / 43

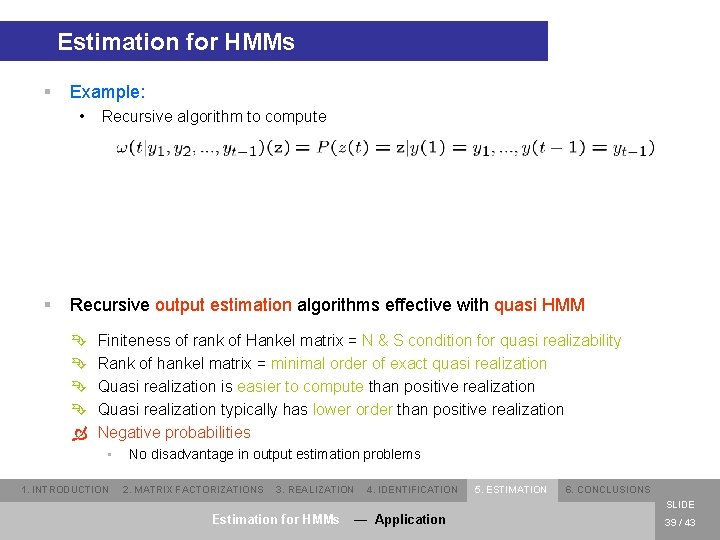
Estimation for HMMs § Example: • § Recursive algorithm to compute Recursive output estimation algorithms effective with quasi HMM Ê Ê Ò Finiteness of rank of Hankel matrix = N & S condition for quasi realizability Rank of hankel matrix = minimal order of exact quasi realization Quasi realization is easier to compute than positive realization Quasi realization typically has lower order than positive realization Negative probabilities • 1. INTRODUCTION No disadvantage in output estimation problems 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Estimation for HMMs — Application 39 / 43
![Finding motifs in DNA sequences Find motifs in muscle specific regulatory regions Zhou Wong Finding motifs in DNA sequences Find motifs in muscle specific regulatory regions [Zhou, Wong]](https://slidetodoc.com/presentation_image_h2/8247431f5aa23f3bd5d36b41cb0b8e5b/image-40.jpg)
Finding motifs in DNA sequences Find motifs in muscle specific regulatory regions [Zhou, Wong] • • Results (compared to results from Motifscanner [Aerts et al. ]) Mef-2 Myf Sp-1 SRF TEF MOTIF PROBABILITY § Make motif model Make quasi background model (see Section realization) Build joint HMM Perform output estimation MOTIF PROBABILITY § POSITION 1. INTRODUCTION 2. MATRIX FACTORIZATIONS POSITION 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE Estimation for HMMs — Application 40 / 43

Conclusions § Two modification to the nonnegative matrix factorization • • § Structured nonnegative matrix factorization Nonnegative matrix factorization without nonnegativity of the factors Two relaxations to the positive realization problem for HMMs • Quasi realization problem • Approximate positive realization problem _ Both methods were applied to modeling DNA sequences § We derive equivalence conditions for HMMs § We propose a new identification method for HMMs _ Method was applied to modeling DNA sequences of HIV virus § Quasi realizations suffice for several estimation problems _ Quasi estimation methods were applied to finding motifs in DNA sequences 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. 6. CONCLUSIONS SLIDE Conclusions — Further research — List of publications 41 / 43

Further research Matrix factorizations § Develop nonnegative matrix factorization with nesting property (cfr. SVD) Hidden Markov models § Investigate Markov models (special case of hidden Markov case) § Develop realization and identification methods that allow to incorporate prior-knowledge in the Markov chain. . . § § Method to estimate minimal order of positive HMM from string probabilities Canonical forms of hidden Markov models Model reduction for hidden Markov models System theory for hidden Markov models with external inputs 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. 6. CONCLUSIONS SLIDE Conclusions — Further research — List of publications 42 / 43

List of publications § § Journal papers • B. Vanluyten, J. C. Willems and B. De Moor. Recursive Filtering using Quasi-Realizations. Lecture Notes in Control and Information Sciences, 341, 367– 374, 2006. • B. Vanluyten, J. C. Willems and B. De Moor. Equivalence of State Representations for Hidden Markov Models. Systems and Control Letters, 57(5), 410– 419, 2008. • B. Vanluyten, J. C. Willems and B. De Moor. Structured Nonnegative Matrix Factorization with Applications to Hidden Markov Realization and Filtering. Accepted for publication in Linear Algebra and its Applications, 2008. • B. Vanluyten, J. C. Willems and B. De Moor. Nonnegative Matrix Factorization without Nonnegativity Constraints on the Factors. Submitted for publication. • B. Vanluyten, J. C. Willems and B. De Moor. Approximate Realization and Estimation for Quasi hidden Markov models. Submitted for publication. International conference papers • I. Goethals, B. Vanluyten, B. De Moor. Reliable spurious mode rejection using self learning algorithms. In Proc. of the International Conference on Modal Analysis Noise and Vibration Engineering (ISMA 2004), Leuven, Belgium, pages 991– 1003, 2004. • B. Vanluyten, J. C. Willems and B. De Moor. Model Reduction of Systems with Symmetries. In Proc. of the 44 th IEEE Conference on Decision and Control (CDC 2005), Seville, Spain, pages 826– 831, 2005. • B. Vanluyten, J. C. Willems and B. De Moor. Matrix Factorization and Stochastic State Representations. In Proc. of the 45 th IEEE Conference on Decision and Control (CDC 2006), San Diego, California, pages 4188 -4193, 2006. • I. Markovsky, J. Boets, B. Vanluyten, K. De Cock, B. De Moor. When is a pole spurious? In Proc. of the International Conference on Noise and Vibration Engineering (ISMA 2007), Leuven, Belgium, pp. 1615– 1626, 2007. • B. Vanluyten, J. C. Willems and B. De Moor. Equivalence of State Representations for Hidden Markov Models. In Proc. of the European Control Conference 2007 (ECC 2007), Kos, Greece, 2007. • B. Vanluyten, J. C. Willems and B. De Moor. A new Approach for the Identification of Hidden Markov Models. In Proc. of the 46 th IEEE Conference on Decision and Control (CDC 2006), New Orleans, Louisiana, 2007. 1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. 6. CONCLUSIONS SLIDE Conclusions — Further research — List of publications 43 / 43

1. INTRODUCTION 2. MATRIX FACTORIZATIONS 3. REALIZATION 4. IDENTIFICATION 5. ESTIMATION 6. CONCLUSIONS SLIDE 44 / 43
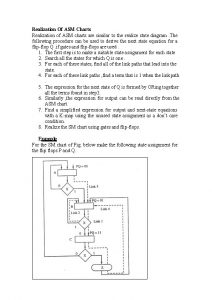
 Asm chart for jk flip flop
Asm chart for jk flip flop Universitei
Universitei Katholieke hogeschool kempen
Katholieke hogeschool kempen Katholieke hogeschool limburg
Katholieke hogeschool limburg Frisdrankautomaat limburg
Frisdrankautomaat limburg Use case realization
Use case realization Cognitive provides various channels for self-realization.
Cognitive provides various channels for self-realization. Type of structure
Type of structure Touching spirit bear questions
Touching spirit bear questions Self realization aristotle
Self realization aristotle Corbiran
Corbiran Cash realization ratio
Cash realization ratio Farzana kite runner
Farzana kite runner What is use case realization
What is use case realization Cash realization ratio
Cash realization ratio Iso 13485 product realization
Iso 13485 product realization Use case realization
Use case realization Leuven
Leuven Cvo landen
Cvo landen Marc ophalvens
Marc ophalvens Zoologisch instituut leuven
Zoologisch instituut leuven Slidetodoc.com
Slidetodoc.com Master actuarial and financial engineering
Master actuarial and financial engineering Smec kuleuven
Smec kuleuven Leo xiii leuven
Leo xiii leuven Kotnet leuven
Kotnet leuven 3c3 hedge fund
3c3 hedge fund Trofozoiet
Trofozoiet Don bosco peda leuven
Don bosco peda leuven International business economics and management
International business economics and management Ku leuven
Ku leuven Exam results ku leuven
Exam results ku leuven Uz leuven antibiotica gids
Uz leuven antibiotica gids Neuromusculair referentiecentrum
Neuromusculair referentiecentrum Open school leuven
Open school leuven Diversiteitsstage
Diversiteitsstage Leuven computer science
Leuven computer science Ku leuven whos who
Ku leuven whos who Ichtus leuven
Ichtus leuven Ku leuven international business economics and management
Ku leuven international business economics and management Ku leuven open access
Ku leuven open access Filip abraham ku leuven
Filip abraham ku leuven Webmail kuleuven be
Webmail kuleuven be Elz leuven noord
Elz leuven noord Ku leuven pret
Ku leuven pret