Oxidative Stress and Repair Oxidative DNA modifications Faiez





- Slides: 27

Oxidative Stress and Repair • Oxidative DNA modifications Faiez Al Nimer • Oxidative lipid modifications Huang Liyue • Oxidative protein modifications Sanyal Nikhilesh • Oxidative DNA and protein damage repair Melanie Neely Willis

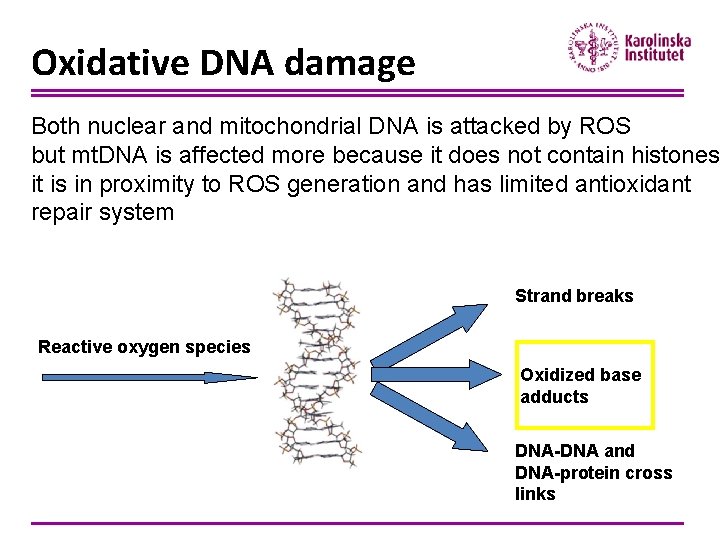
Oxidative DNA damage Both nuclear and mitochondrial DNA is attacked by ROS but mt. DNA is affected more because it does not contain histones it is in proximity to ROS generation and has limited antioxidant repair system Strand breaks Reactive oxygen species Oxidized base adducts DNA-DNA and DNA-protein cross links

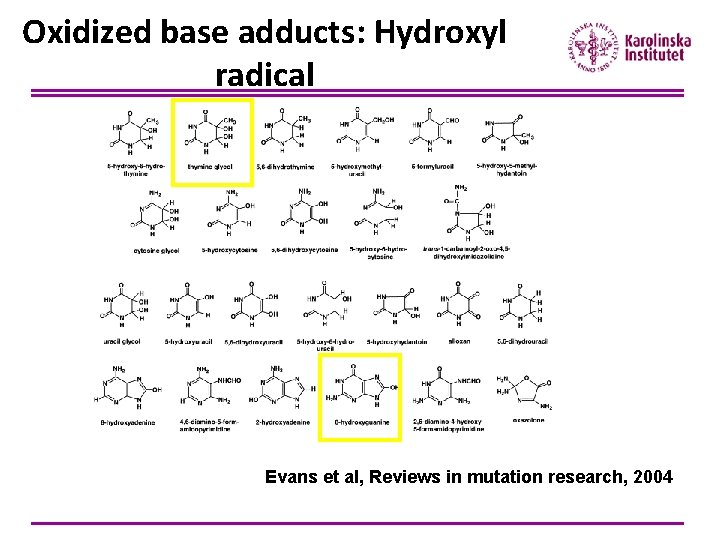
Oxidized base adducts: Hydroxyl radical Evans et al, Reviews in mutation research, 2004

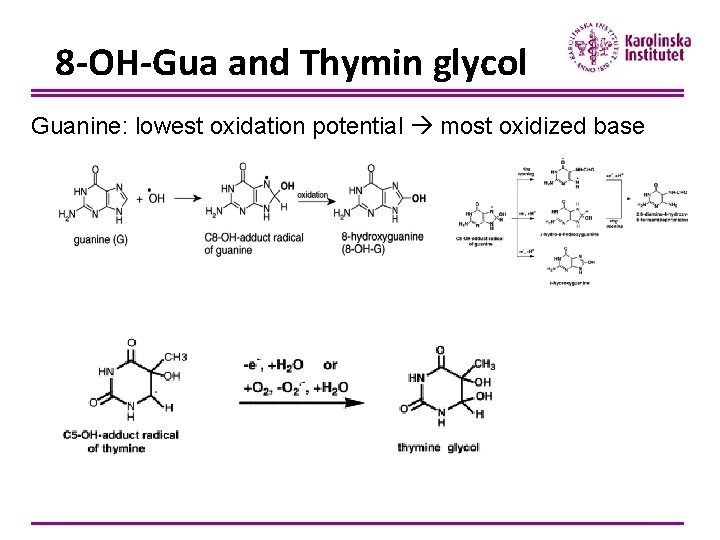
8 -OH-Gua and Thymin glycol Guanine: lowest oxidation potential most oxidized base

Cell viability and disease ØFor 8 -OH-Gua: Results in C→A and G→T transversions, leading to mutations Affects transcription factor binding and gene expression Interferes with methylation of i. e. oncogenes ØFor thymin glycol: T→C transitions have been noted Blocks replication

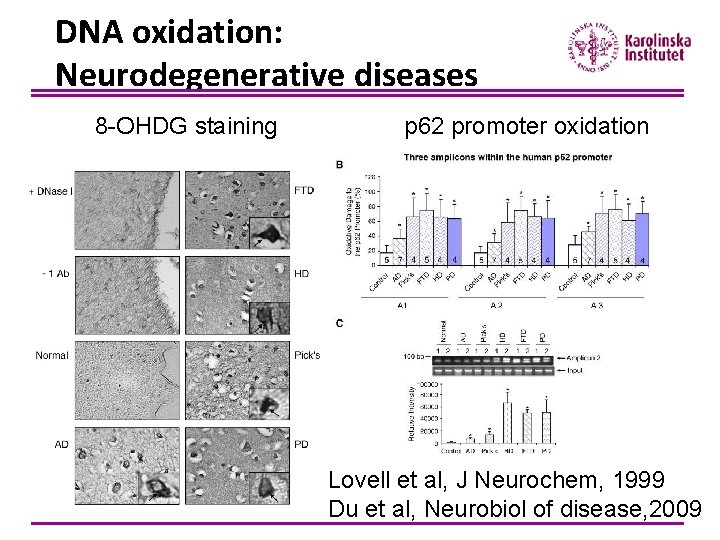
DNA oxidation: Neurodegenerative diseases 8 -OHDG staining p 62 promoter oxidation Lovell et al, J Neurochem, 1999 Du et al, Neurobiol of disease, 2009

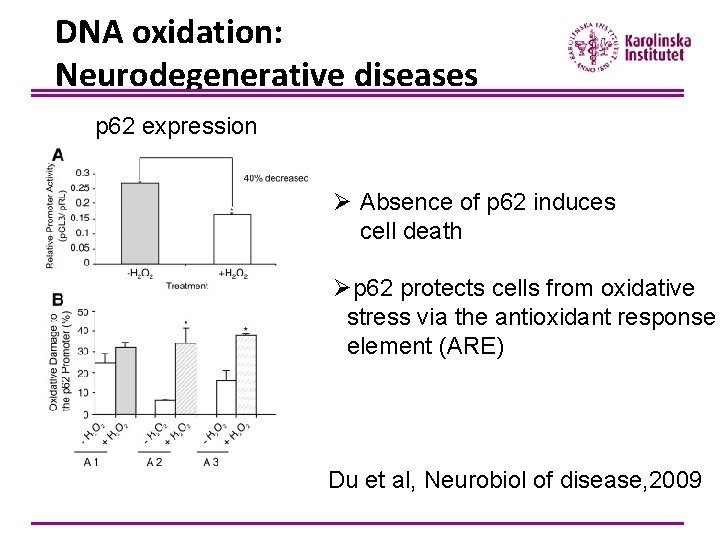
DNA oxidation: Neurodegenerative diseases p 62 expression Ø Absence of p 62 induces cell death Øp 62 protects cells from oxidative stress via the antioxidant response element (ARE) Du et al, Neurobiol of disease, 2009

Oxidative Stress and Repair • Oxidative DNA modifications Faiez Al Nimer • Oxidative lipid modifications Huang Liyue • Oxidative protein modifications Sanyal Nikhilesh • Oxidative DNA and protein damage repair Melanie Neely Willis

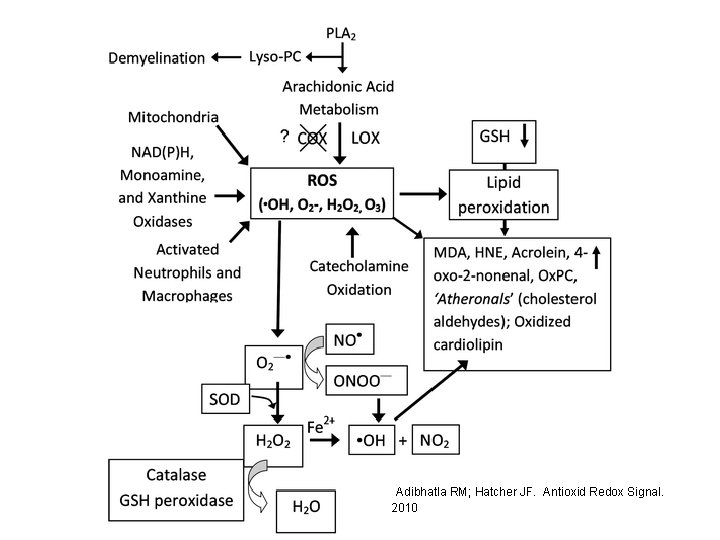
Lipid modifications by reactive oxygen species (ROS) • Overproduction of ROS damages cellular components, including lipids, leading to decline in physiological function and cell death

Reaction of ROS with lipids lead to • Lipid oxidation • Lipid peroxidation

Lipid Oxidation • Initiation – A radical is formed • Propagation – Radicals react and transfer their unpaired electron to other compounds • Termination – Two radicals combine to stop the reaction

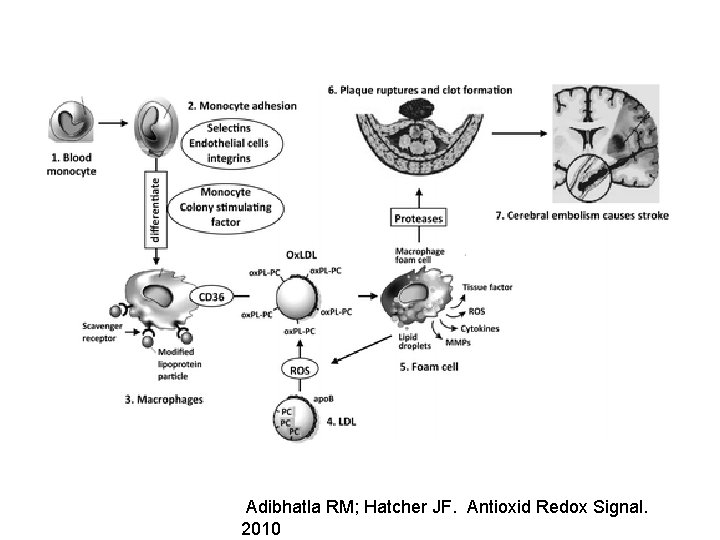
Adibhatla RM; Hatcher JF. Antioxid Redox Signal. 2010

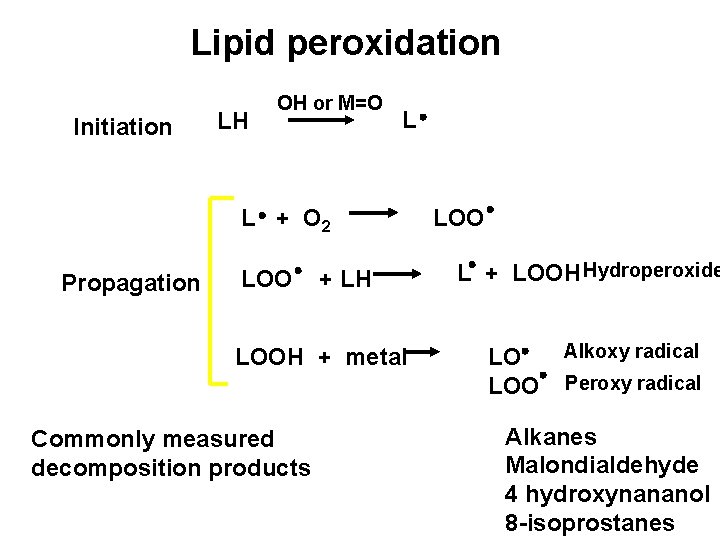
Lipid peroxidation Initiation LH OH or M=O L L + O 2 Propagation LOO + LH LOOH + metal Commonly measured decomposition products LOO L + LOOH Hydroperoxide LO LOO Alkoxy radical Peroxy radical Alkanes Malondialdehyde 4 hydroxynananol 8 -isoprostanes

Adibhatla RM; Hatcher JF. Antioxid Redox Signal. 2010

Oxidative Stress and Repair • Oxidative DNA modifications Faiez Al Nimer • Oxidative lipid modifications Huang Liyue • Oxidative protein modifications Sanyal Nikhilesh • Oxidative DNA and protein damage repair Melanie Neely Willis

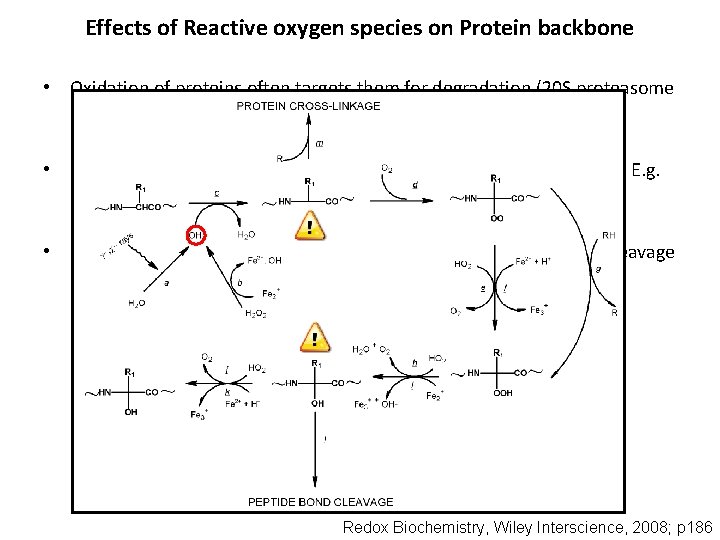
Effects of Reactive oxygen species on Protein backbone • Oxidation of proteins often targets them for degradation (20 S proteasome and other proteases). • Several age-related disorders are characterised by Protein oxidation. E. g. Parkinson’s disease, Amyotropic Lateral sclerosis. • Product of the oxidation (Peptide alkoxyl radical) is susceptible to cleavage by diamide or a-amidation pathways. Redox Biochemistry, Wiley Interscience, 2008; p 186

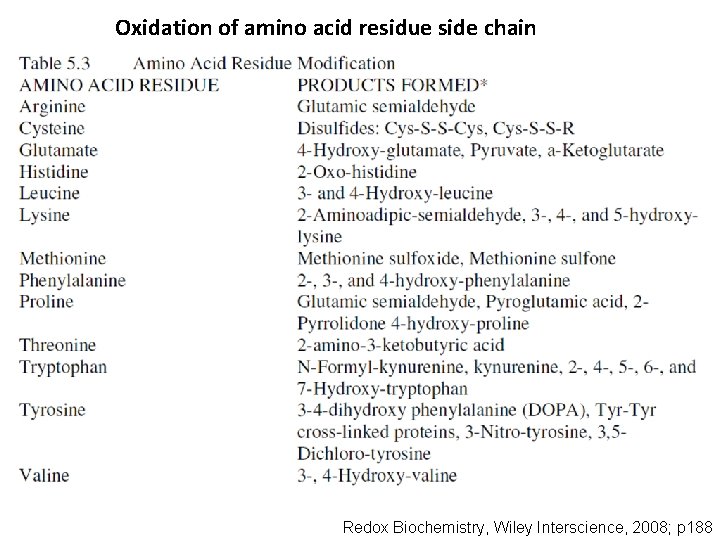
Oxidation of amino acid residue side chain Redox Biochemistry, Wiley Interscience, 2008; p 188

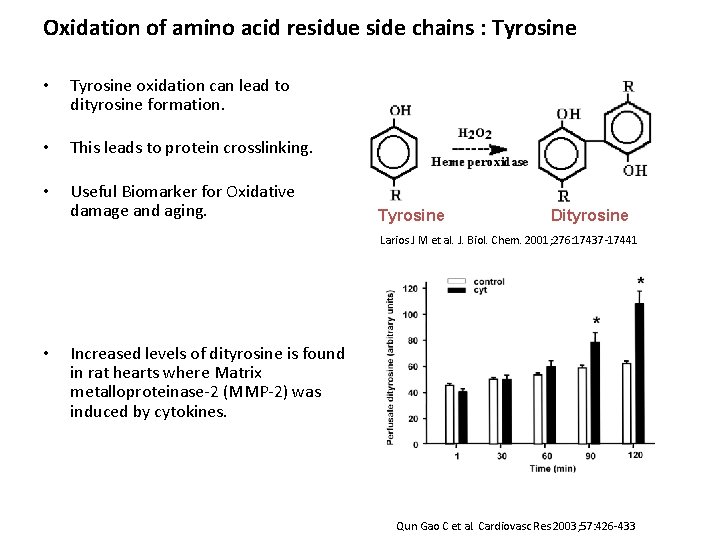
Oxidation of amino acid residue side chains : Tyrosine • Tyrosine oxidation can lead to dityrosine formation. • This leads to protein crosslinking. • Useful Biomarker for Oxidative damage and aging. Tyrosine Dityrosine Larios J M et al. J. Biol. Chem. 2001; 276: 17437 -17441 • Increased levels of dityrosine is found in rat hearts where Matrix metalloproteinase-2 (MMP-2) was induced by cytokines. Qun Gao C et al. Cardiovasc Res 2003; 57: 426 -433

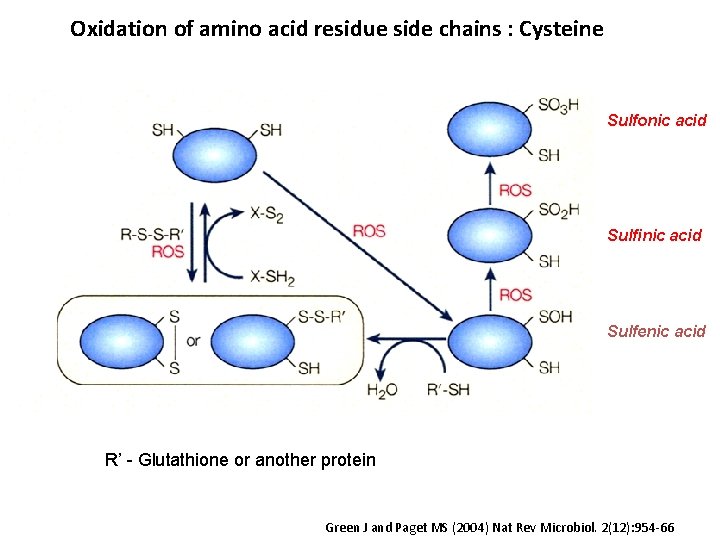
Oxidation of amino acid residue side chains : Cysteine Sulfonic acid Sulfinic acid Sulfenic acid R’ - Glutathione or another protein Green J and Paget MS (2004) Nat Rev Microbiol. 2(12): 954 -66

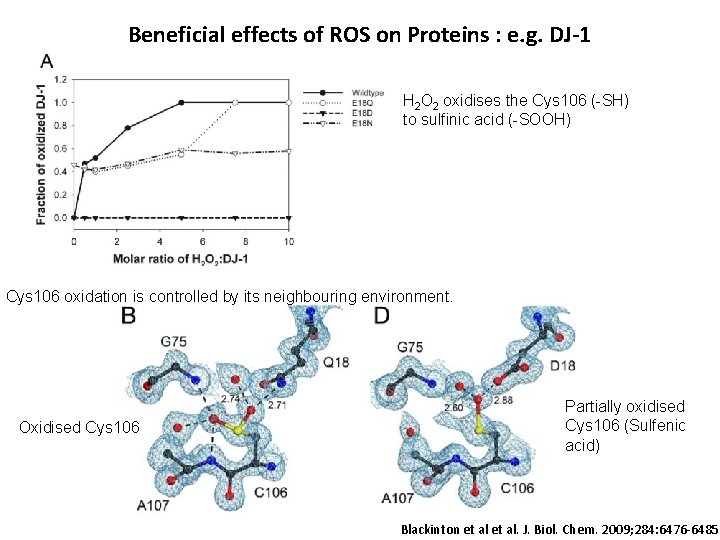
Beneficial effects of ROS on Proteins : e. g. DJ-1 H 2 O 2 oxidises the Cys 106 (-SH) to sulfinic acid (-SOOH) Cys 106 oxidation is controlled by its neighbouring environment. Oxidised Cys 106 Partially oxidised Cys 106 (Sulfenic acid) Blackinton et al. J. Biol. Chem. 2009; 284: 6476 -6485

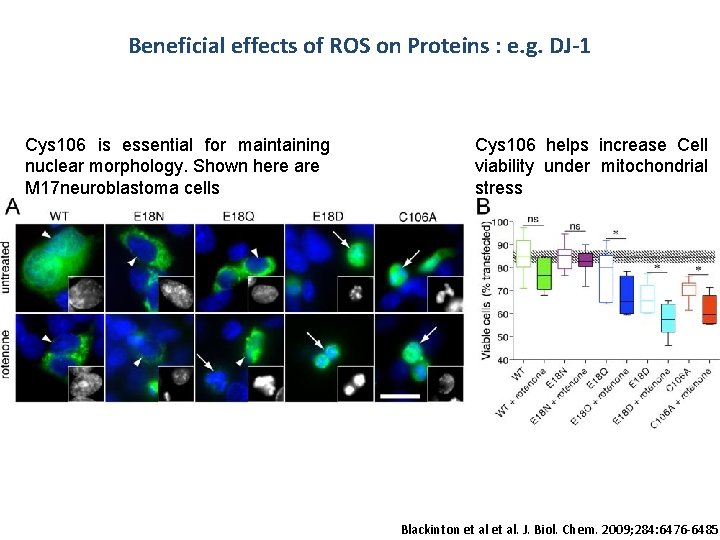
Beneficial effects of ROS on Proteins : e. g. DJ-1 Cys 106 is essential for maintaining nuclear morphology. Shown here are M 17 neuroblastoma cells Cys 106 helps increase Cell viability under mitochondrial stress Blackinton et al. J. Biol. Chem. 2009; 284: 6476 -6485

Oxidative Stress and Repair • Oxidative DNA modifications Faiez Al Nimer • Oxidative lipid modifications Huang Liyue • Oxidative protein modifications Sanyal Nikhilesh • Oxidative DNA and protein damage repair Melanie Neely Willis

DNA & Protein Repair Enzymes Oxidation of pyrimidine and purine bases leads to DNA damage. Oxidation of methionine and cysteine residues can lead to protein damage and aggregation. DNA damage is repaired by several enzymes in the Base Excision Repair pathway (BER). Proteins are repaired by methionine sulfoxide reductases (Msr. A, Msr. B) and sulfiredoxins.

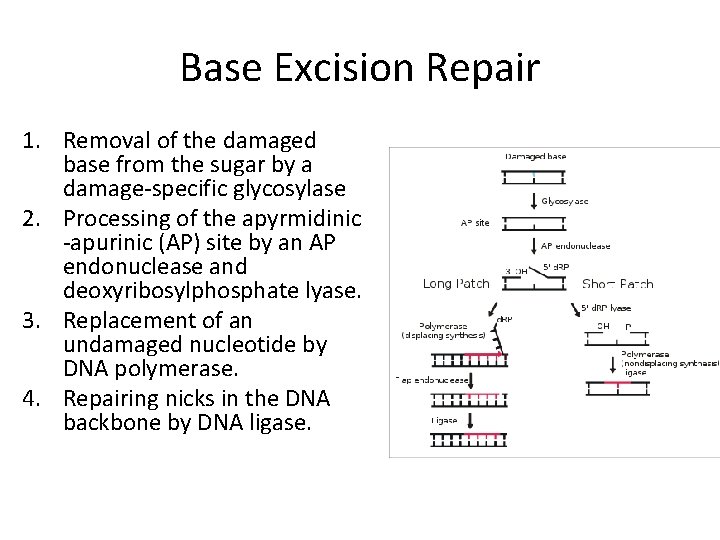
Base Excision Repair 1. Removal of the damaged base from the sugar by a damage-specific glycosylase 2. Processing of the apyrmidinic -apurinic (AP) site by an AP endonuclease and deoxyribosylphosphate lyase. 3. Replacement of an undamaged nucleotide by DNA polymerase. 4. Repairing nicks in the DNA backbone by DNA ligase.

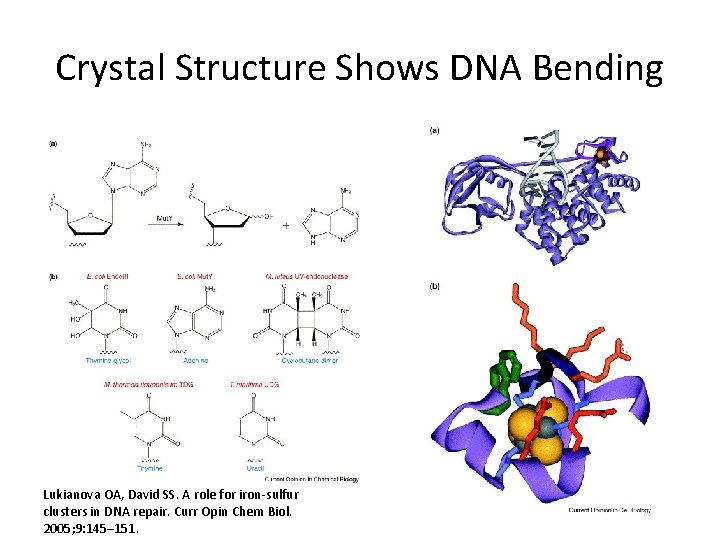
Crystal Structure Shows DNA Bending Lukianova OA, David SS. A role for iron-sulfur clusters in DNA repair. Curr Opin Chem Biol. 2005; 9: 145– 151.

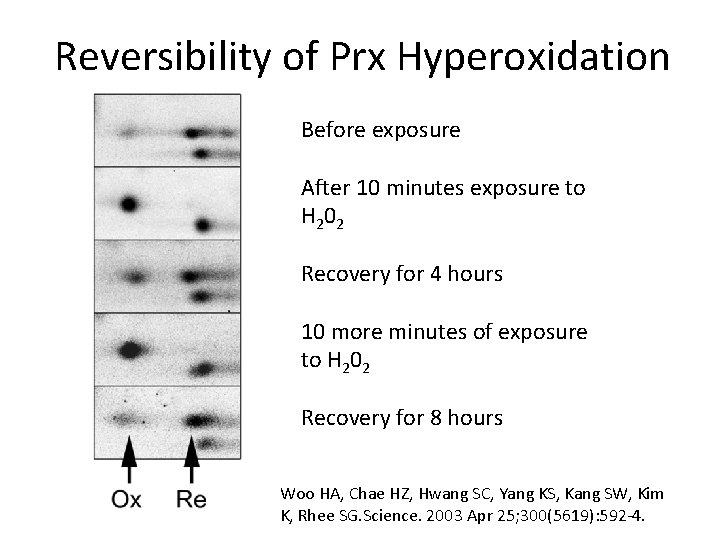
Reversibility of Prx Hyperoxidation Before exposure After 10 minutes exposure to H 2 0 2 Recovery for 4 hours 10 more minutes of exposure to H 202 Recovery for 8 hours Woo HA, Chae HZ, Hwang SC, Yang KS, Kang SW, Kim K, Rhee SG. Science. 2003 Apr 25; 300(5619): 592 -4.

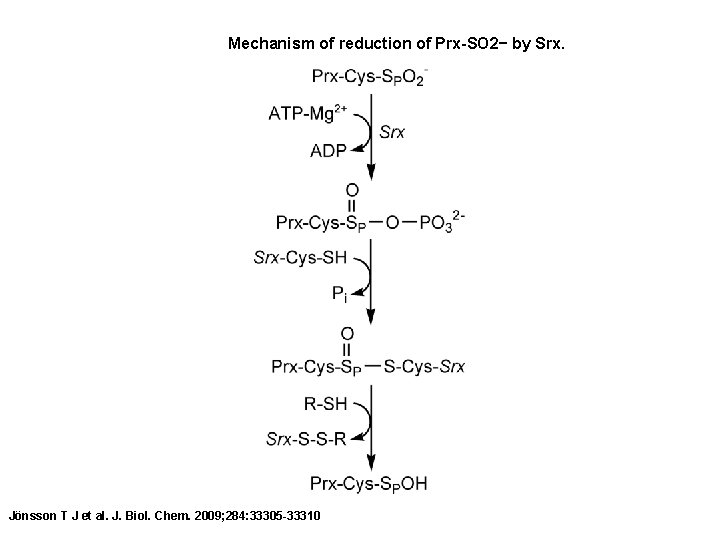
Mechanism of reduction of Prx-SO 2− by Srx. Jönsson T J et al. J. Biol. Chem. 2009; 284: 33305 -33310
 Faiez al nimer
Faiez al nimer Excision repair
Excision repair Base excision repair vs mismatch repair
Base excision repair vs mismatch repair Dna polymerase
Dna polymerase Plant modifications of roots stems and leaves
Plant modifications of roots stems and leaves Accommodations and modifications
Accommodations and modifications Difference in accommodations and modifications
Difference in accommodations and modifications Difference in accommodations and modifications
Difference in accommodations and modifications True stress vs engineering stress
True stress vs engineering stress Chapter 10 stress responses and stress management
Chapter 10 stress responses and stress management Coding dna and non coding dna
Coding dna and non coding dna Blackhawk king air 350
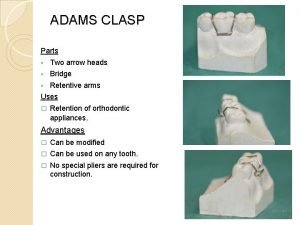
Blackhawk king air 350 Adams clasp making
Adams clasp making Discours direct
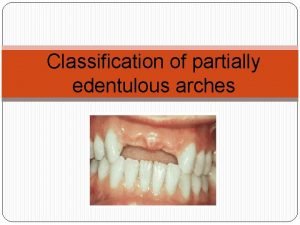
Discours direct Class 5 kennedy classification
Class 5 kennedy classification Accommodation vs modification
Accommodation vs modification Definition of axial stress
Definition of axial stress Replication
Replication Bioflix activity dna replication dna replication diagram
Bioflix activity dna replication dna replication diagram What role does dna polymerase play in copying dna?
What role does dna polymerase play in copying dna? Chapter 11 dna and genes
Chapter 11 dna and genes Amino urea
Amino urea Transamination and oxidative deamination
Transamination and oxidative deamination Transamination and oxidative deamination
Transamination and oxidative deamination Biliverdin color
Biliverdin color Oxidative deamination of amino acids
Oxidative deamination of amino acids Fructose breakdown pathway
Fructose breakdown pathway Uncouple oxidative phosphorylation
Uncouple oxidative phosphorylation