Oxidative Damage of DNA Oxidative damage results from













- Slides: 43

Oxidative Damage of DNA Oxidative damage results from aerobic metabolism, environmental toxins, activated macrophages, and signaling molecules (NO) Compartmentation limits oxidative DNA damage

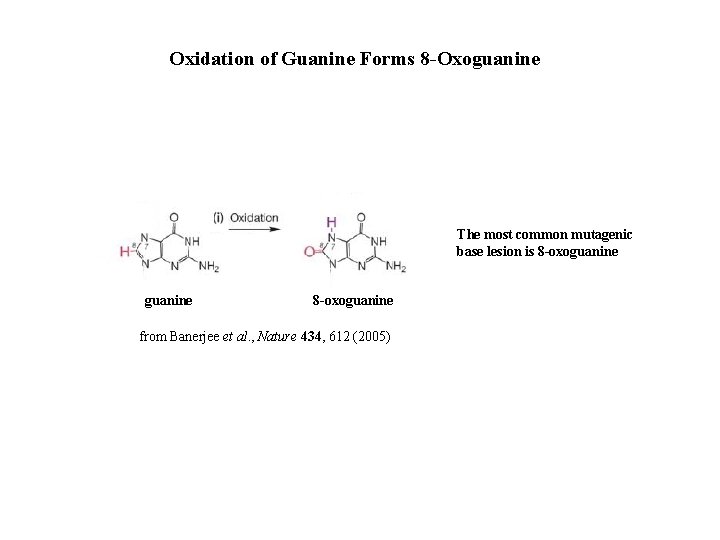
Oxidation of Guanine Forms 8 -Oxoguanine The most common mutagenic base lesion is 8 -oxoguanine from Banerjee et al. , Nature 434, 612 (2005)

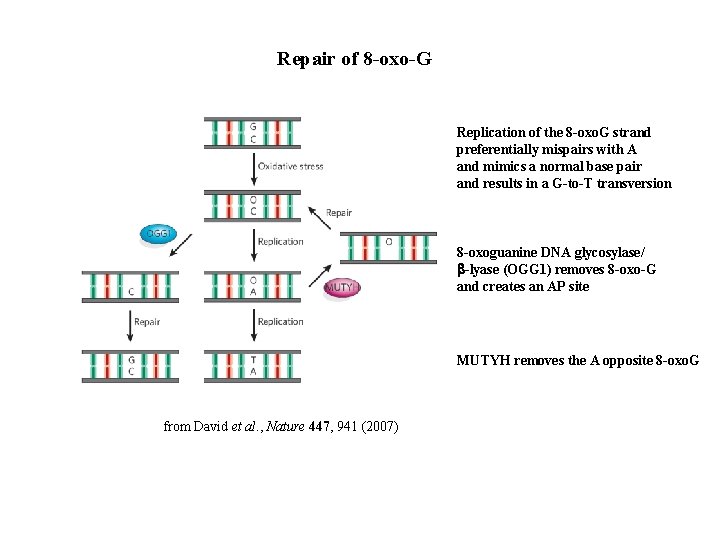
Repair of 8 -oxo-G Replication of the 8 -oxo. G strand preferentially mispairs with A and mimics a normal base pair and results in a G-to-T transversion 8 -oxoguanine DNA glycosylase/ b-lyase (OGG 1) removes 8 -oxo-G and creates an AP site MUTYH removes the A opposite 8 -oxo. G from David et al. , Nature 447, 941 (2007)

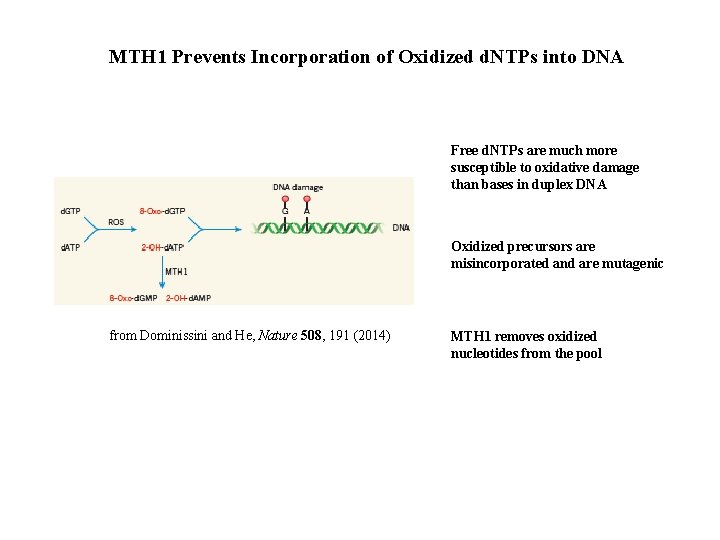
MTH 1 Prevents Incorporation of Oxidized d. NTPs into DNA Free d. NTPs are much more susceptible to oxidative damage than bases in duplex DNA Oxidized precursors are misincorporated and are mutagenic from Dominissini and He, Nature 508, 191 (2014) MTH 1 removes oxidized nucleotides from the pool

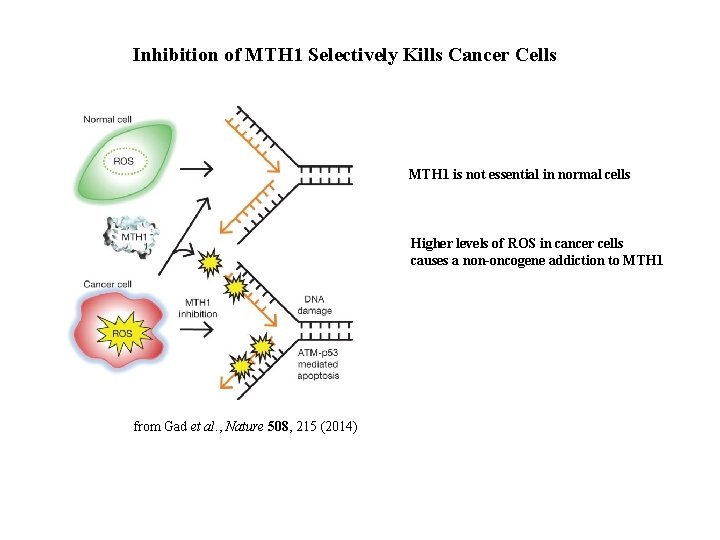
Inhibition of MTH 1 Selectively Kills Cancer Cells MTH 1 is not essential in normal cells Higher levels of ROS in cancer cells causes a non-oncogene addiction to MTH 1 from Gad et al. , Nature 508, 215 (2014)

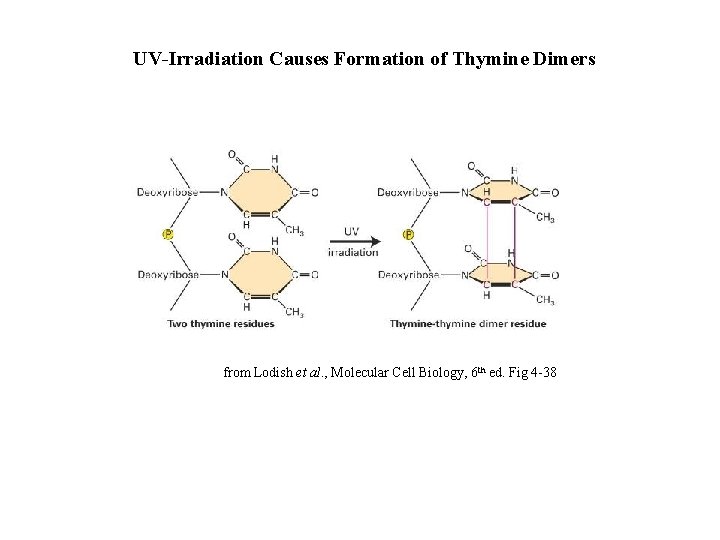
UV-Irradiation Causes Formation of Thymine Dimers from Lodish et al. , Molecular Cell Biology, 6 th ed. Fig 4 -38

Nonenzymatic Methylation of DNA Formation of 600 3 -me-A residues/cell/day are caused by S-adenosylmethionine 3 -me-A is cytotoxic and is repaired by 3 -me-A-DNA glycosylase 7 -me-G is the main aberrant base present in DNA and is repaired by nonenzymatic cleavage of the glycosyl bond

Effect of Chemical Mutagens Nitrous acid causes deamination of C to U and A to HX U base pairs with A HX base pairs with C

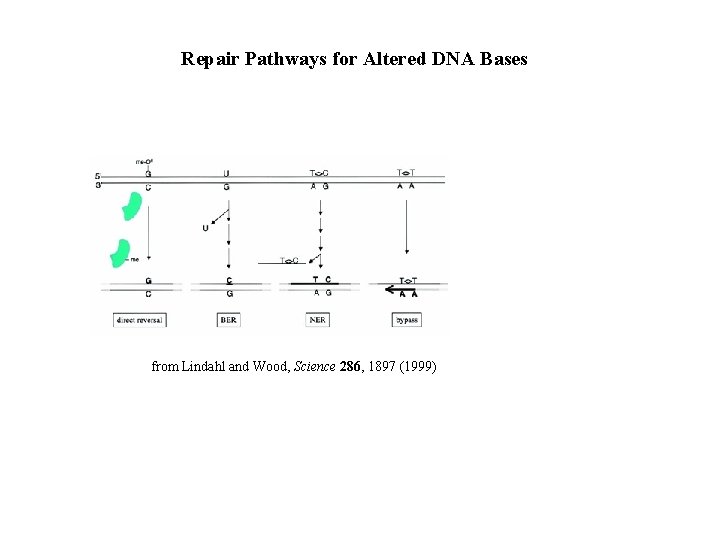
Repair Pathways for Altered DNA Bases from Lindahl and Wood, Science 286, 1897 (1999)

Direct Repair of DNA Photoreactivation of pyrimidine dimers by photolyase restores the original DNA structure O 6 -methylguanine is repaired by removal of methyl group by MGMT 1 -methyladenine and 3 -methylcytosine are repaired by oxidative demethylation

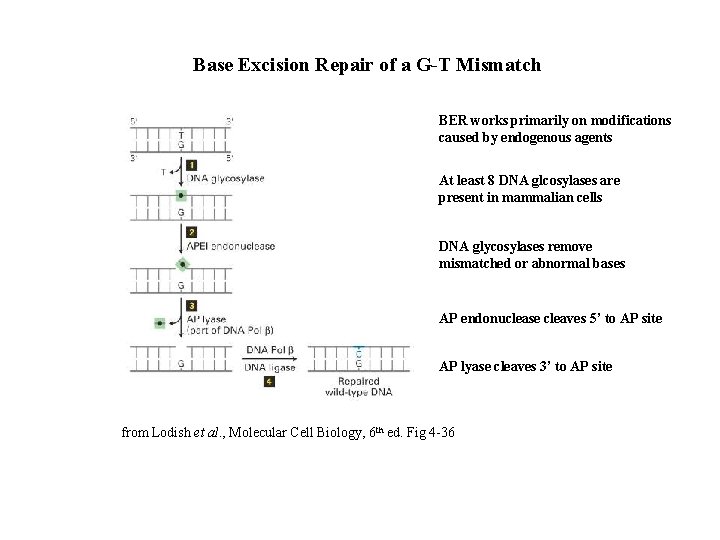
Base Excision Repair of a G-T Mismatch BER works primarily on modifications caused by endogenous agents At least 8 DNA glcosylases are present in mammalian cells DNA glycosylases remove mismatched or abnormal bases AP endonuclease cleaves 5’ to AP site AP lyase cleaves 3’ to AP site from Lodish et al. , Molecular Cell Biology, 6 th ed. Fig 4 -36

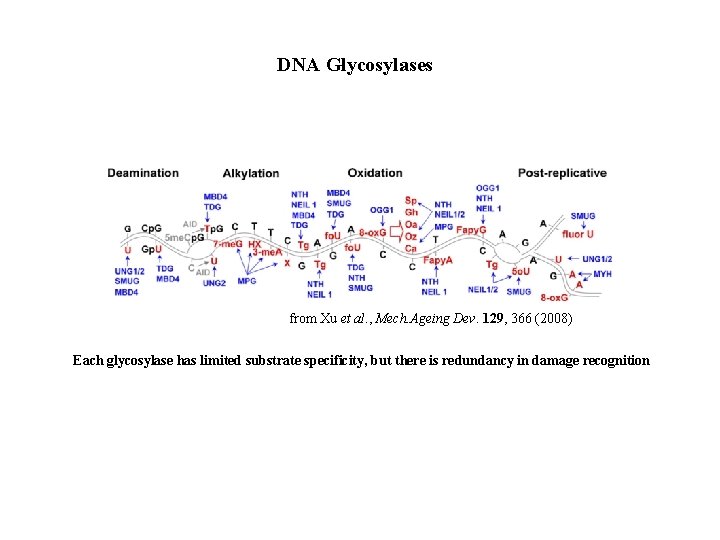
DNA Glycosylases from Xu et al. , Mech. Ageing Dev. 129, 366 (2008) Each glycosylase has limited substrate specificity, but there is redundancy in damage recognition

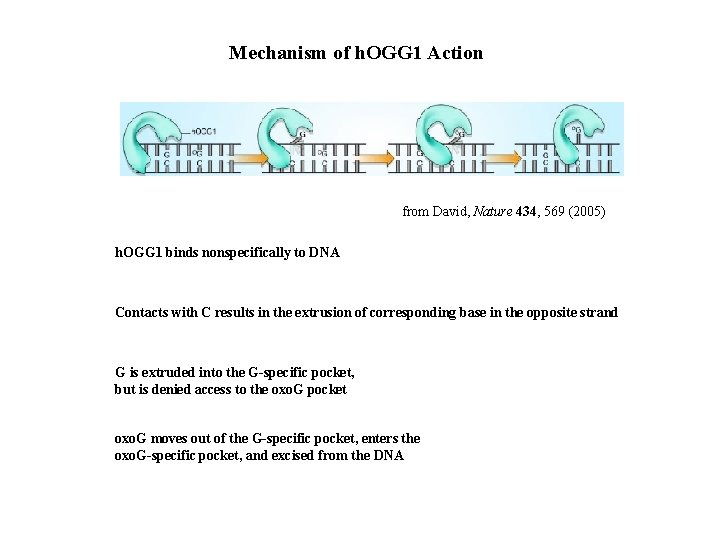
Mechanism of h. OGG 1 Action from David, Nature 434, 569 (2005) h. OGG 1 binds nonspecifically to DNA Contacts with C results in the extrusion of corresponding base in the opposite strand G is extruded into the G-specific pocket, but is denied access to the oxo. G pocket oxo. G moves out of the G-specific pocket, enters the oxo. G-specific pocket, and excised from the DNA

Nucleotide Excision Repairs helix-distorting lesions UV-induced pyrimidine dimers Bulky adducts Intrastrand crosslinks ROS-generated cyclopurines

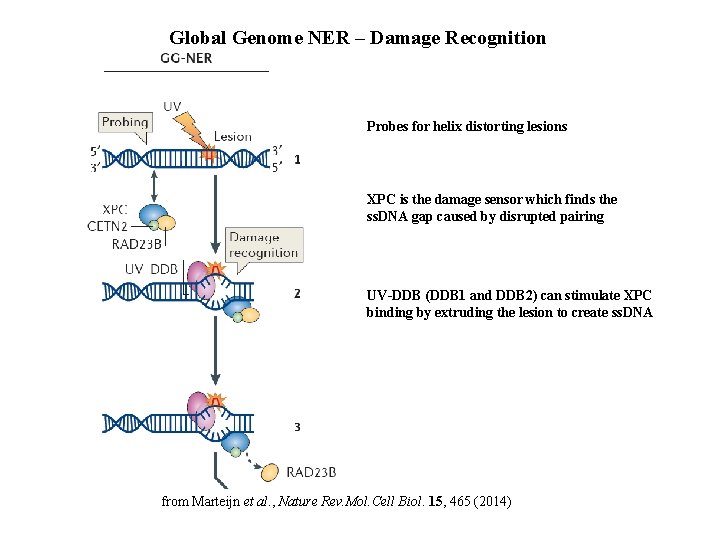
Global Genome NER – Damage Recognition Probes for helix distorting lesions XPC is the damage sensor which finds the ss. DNA gap caused by disrupted pairing UV-DDB (DDB 1 and DDB 2) can stimulate XPC binding by extruding the lesion to create ss. DNA from Marteijn et al. , Nature Rev. Mol. Cell Biol. 15, 465 (2014)

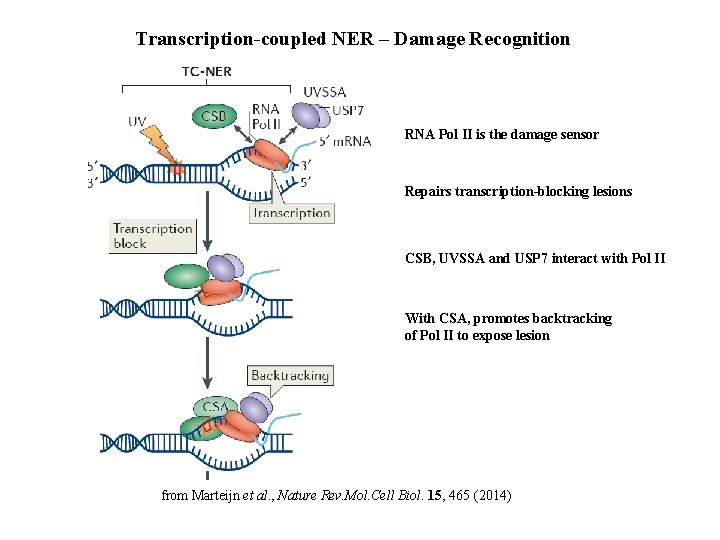
Transcription-coupled NER – Damage Recognition RNA Pol II is the damage sensor Repairs transcription-blocking lesions CSB, UVSSA and USP 7 interact with Pol II With CSA, promotes backtracking of Pol II to expose lesion from Marteijn et al. , Nature Rev. Mol. Cell Biol. 15, 465 (2014)

NER – Lesion Verification TFIIH complex is recruited to the lesion XPB and XPD are helicases with opposite polarity XPD verifies the existence of lesions and XPA binds to altered nucleotides RPA protects the undamaged strand from nucleases XPG nuclease binds to the complex from Marteijn et al. , Nature Rev. Mol. Cell Biol. 15, 465 (2014)

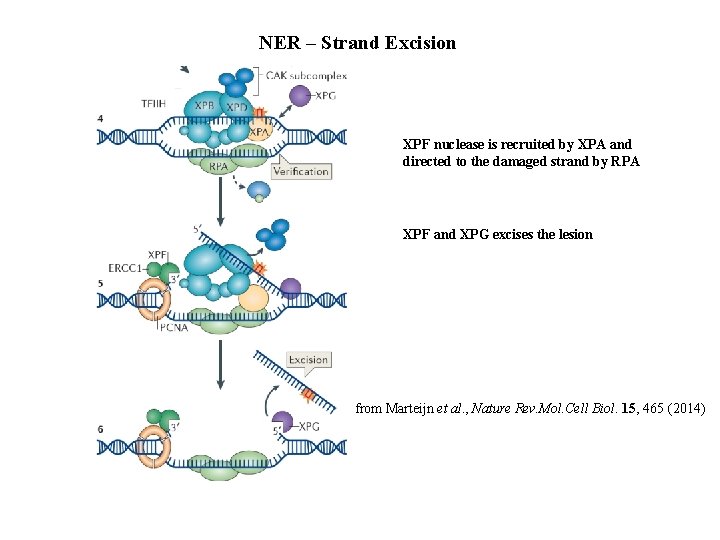
NER – Strand Excision XPF nuclease is recruited by XPA and directed to the damaged strand by RPA XPF and XPG excises the lesion from Marteijn et al. , Nature Rev. Mol. Cell Biol. 15, 465 (2014)

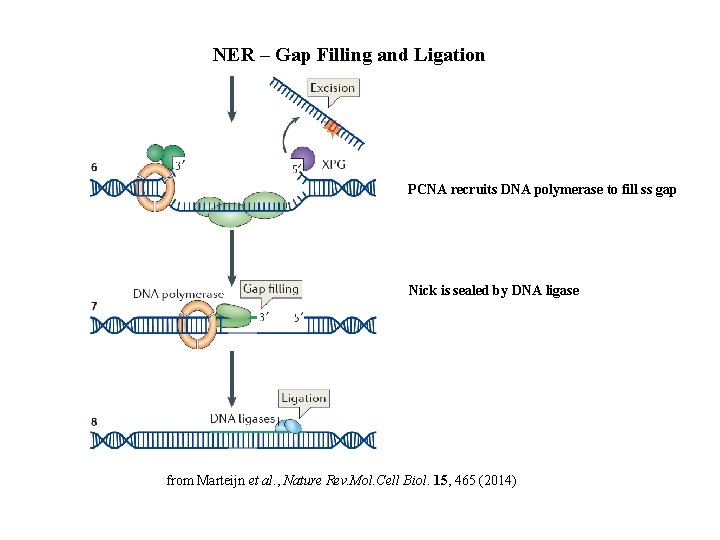
NER – Gap Filling and Ligation PCNA recruits DNA polymerase to fill ss gap Nick is sealed by DNA ligase from Marteijn et al. , Nature Rev. Mol. Cell Biol. 15, 465 (2014)

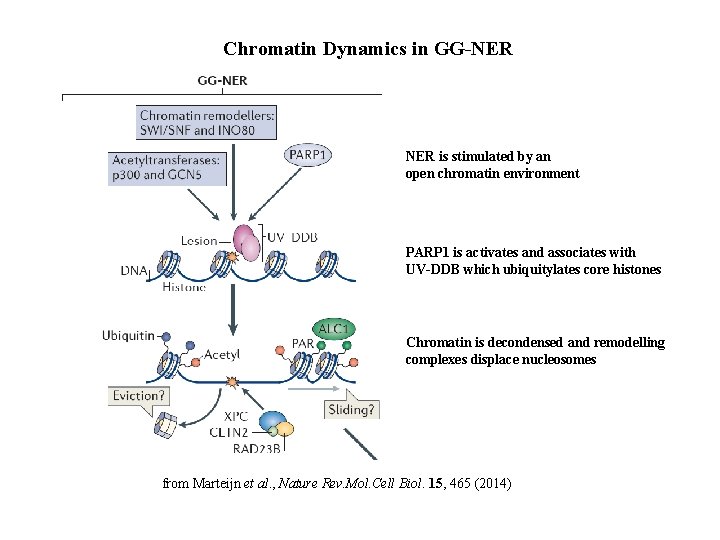
Chromatin Dynamics in GG-NER is stimulated by an open chromatin environment PARP 1 is activates and associates with UV-DDB which ubiquitylates core histones Chromatin is decondensed and remodelling complexes displace nucleosomes from Marteijn et al. , Nature Rev. Mol. Cell Biol. 15, 465 (2014)

Clinical Implications of Defective NER GG-NER is elevated in germ cells to maintain the entire genome to prevent mutagenesis Defective GG-NER increases cancer predisposition Xeroderma pigmentosum TC-NER is elevated in somatic cells to repair expressed genes to prevent cell death Defective TC-NER causes premature cell death, neurodegeration and accelerates aging Cockayne Syndrome

Mismatch Repairs DNA replication errors and insertion-deletion loops Decreases mutation frequency by 102 - 103 Plays a role in triplet repeat expansion, somatic hypermutation and class switch recombination

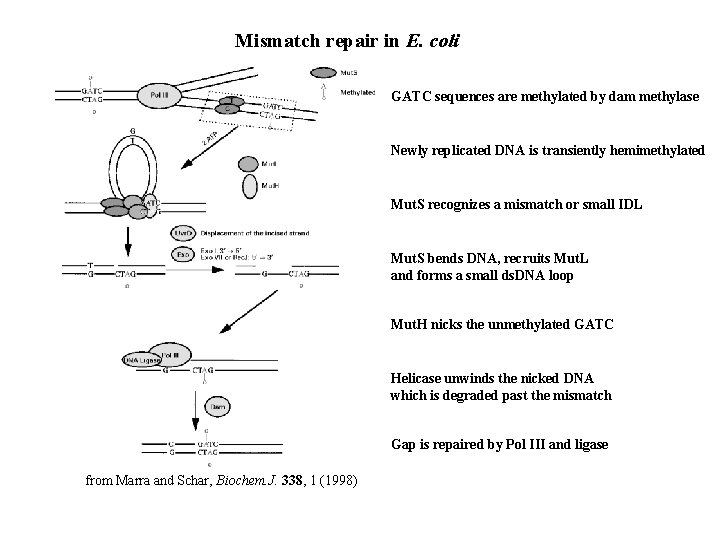
Mismatch repair in E. coli GATC sequences are methylated by dam methylase Newly replicated DNA is transiently hemimethylated Mut. S recognizes a mismatch or small IDL Mut. S bends DNA, recruits Mut. L and forms a small ds. DNA loop Mut. H nicks the unmethylated GATC Helicase unwinds the nicked DNA which is degraded past the mismatch Gap is repaired by Pol III and ligase from Marra and Schar, Biochem. J. 338, 1 (1998)

Mismatch Repair in Eukaryotes Mut. S homologs recognize mismatch and form a ternary complex with Mul. L homologs and the mismatch PMS 2 is a mismatch-activated strandspecific nuclease, and the break is directed to the strand containing nicks Nicks are provided by the ribonucleotide excision repair pathway EXO 1 excises the mismatch and the gap is filled in by PCNA, Pold and DNA ligase Defective mismatch repair is the primary cause of certain types of human cancers from Hsieh and Yamane, Mech. Ageing Dev. 129, 391 (2008)

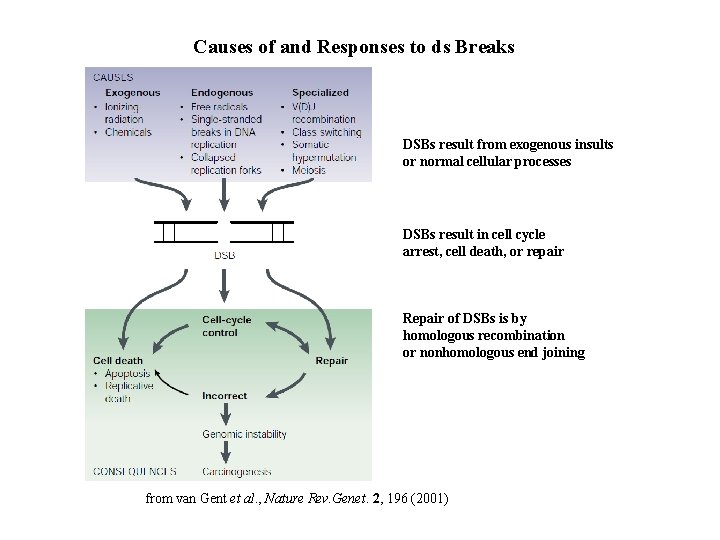
Causes of and Responses to ds Breaks DSBs result from exogenous insults or normal cellular processes DSBs result in cell cycle arrest, cell death, or repair Repair of DSBs is by homologous recombination or nonhomologous end joining from van Gent et al. , Nature Rev. Genet. 2, 196 (2001)

Kinases are Recruited to Sites of DNA Damage Some substrates are shared between these structurally-related kinases DNA-PK promotes NHEJ ATM promotes HR and NHEJ ATR promotes cell cycle arrest and is involved in DNA replication stress responses from Blackford and Jackson, Molecular Cell 66, 801 (2017)

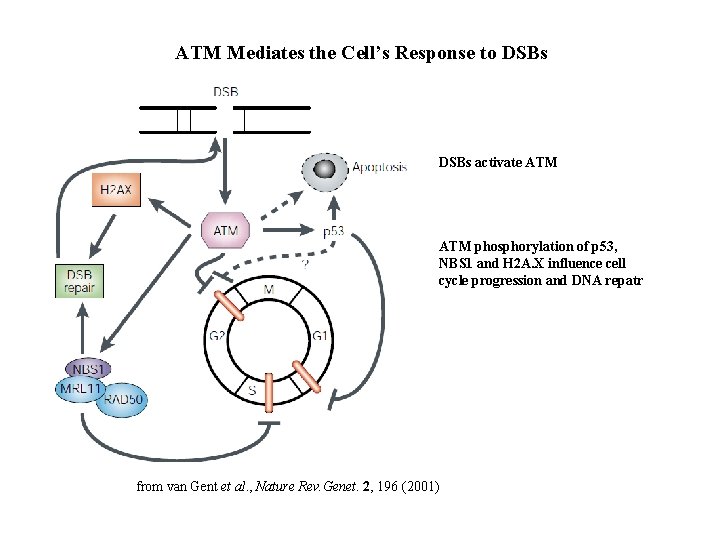
ATM Mediates the Cell’s Response to DSBs activate ATM phosphorylation of p 53, NBS 1 and H 2 A. X influence cell cycle progression and DNA repatr from van Gent et al. , Nature Rev. Genet. 2, 196 (2001)

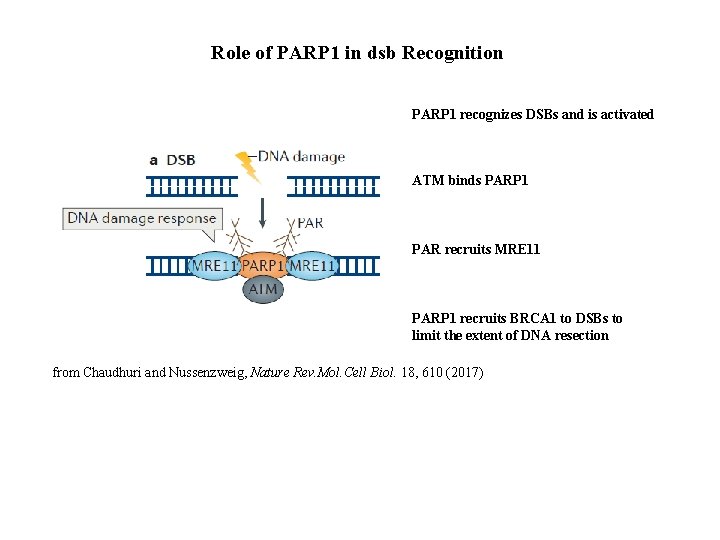
Role of PARP 1 in dsb Recognition PARP 1 recognizes DSBs and is activated ATM binds PARP 1 PAR recruits MRE 11 PARP 1 recruits BRCA 1 to DSBs to limit the extent of DNA resection from Chaudhuri and Nussenzweig, Nature Rev. Mol. Cell Biol. 18, 610 (2017)

Chromatin Modification at DSB from Panier and Boulton, Nature Rev. Mol. Cell Biol. 15, 7 (2014) MRN recruits ATM which phosphorylates H 2 A. X g. H 2 A. X is recognized by MDC 1 which recruits more MRN and ATM phosphorylates MDC 1 which recruits RNF 8 ubiquitylates H 1 which recruits RNF 168 ubiquitylates H 2 A 53 BP 1 is recruited by H 4 K 20 me 2 and H 2 AK 13/15 Ub

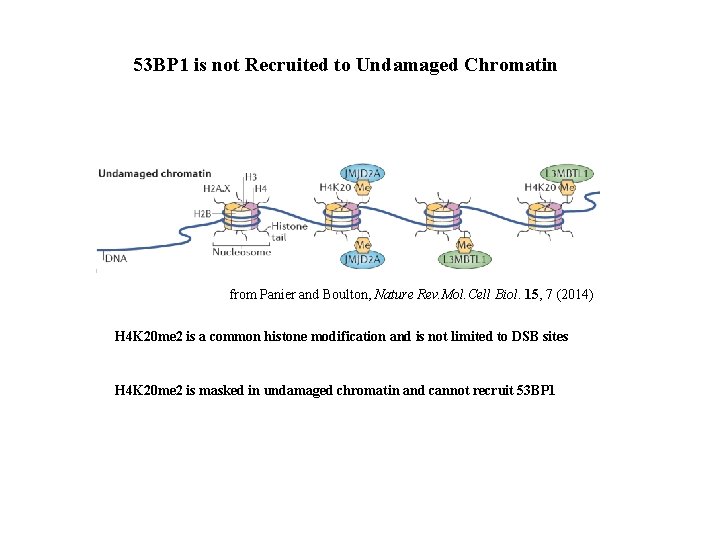
53 BP 1 is not Recruited to Undamaged Chromatin from Panier and Boulton, Nature Rev. Mol. Cell Biol. 15, 7 (2014) H 4 K 20 me 2 is a common histone modification and is not limited to DSB sites H 4 K 20 me 2 is masked in undamaged chromatin and cannot recruit 53 BP 1

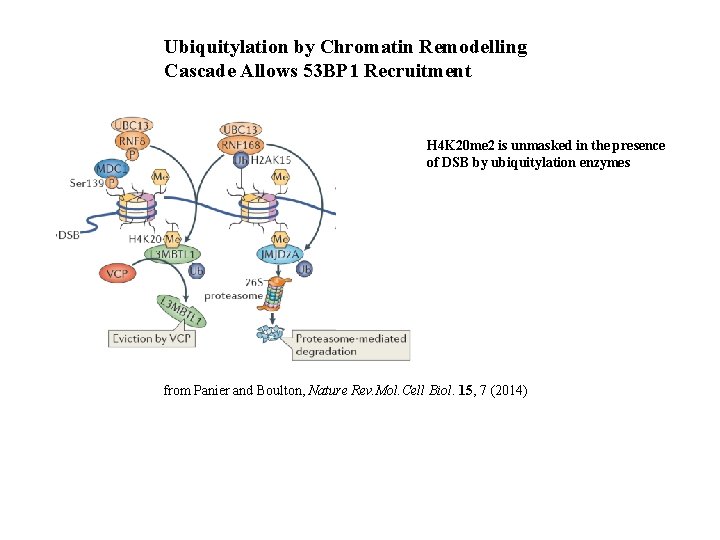
Ubiquitylation by Chromatin Remodelling Cascade Allows 53 BP 1 Recruitment H 4 K 20 me 2 is unmasked in the presence of DSB by ubiquitylation enzymes from Panier and Boulton, Nature Rev. Mol. Cell Biol. 15, 7 (2014)

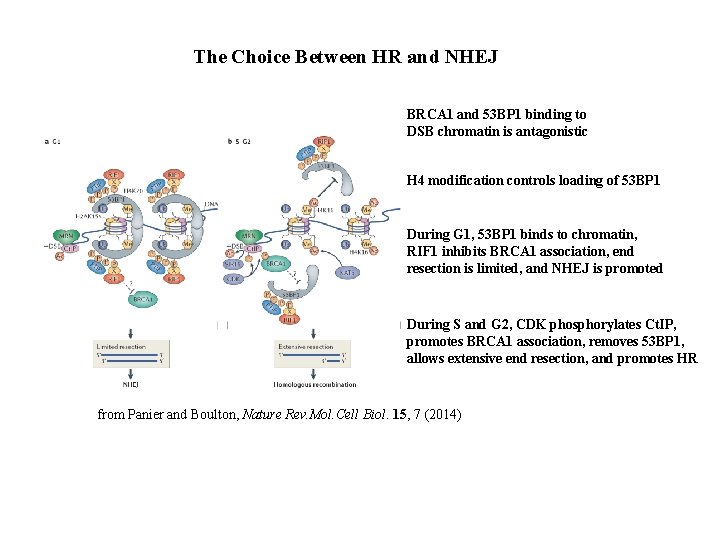
The Choice Between HR and NHEJ BRCA 1 and 53 BP 1 binding to DSB chromatin is antagonistic H 4 modification controls loading of 53 BP 1 During G 1, 53 BP 1 binds to chromatin, RIF 1 inhibits BRCA 1 association, end resection is limited, and NHEJ is promoted During S and G 2, CDK phosphorylates Ct. IP, promotes BRCA 1 association, removes 53 BP 1, allows extensive end resection, and promotes HR from Panier and Boulton, Nature Rev. Mol. Cell Biol. 15, 7 (2014)

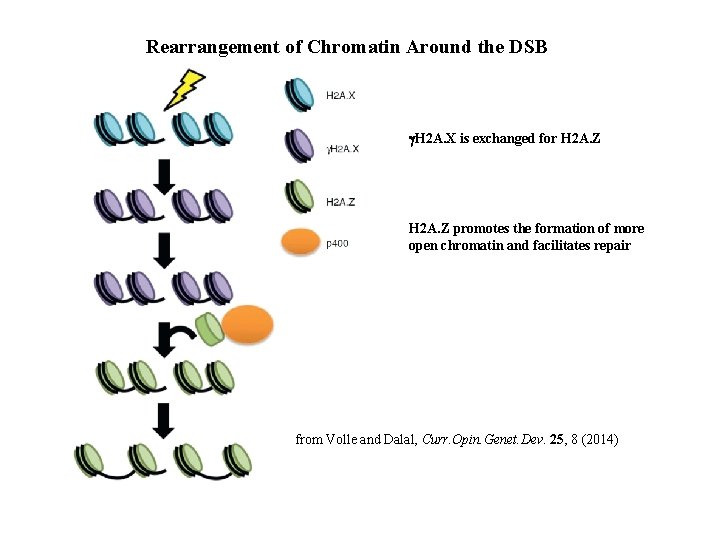
Rearrangement of Chromatin Around the DSB g. H 2 A. X is exchanged for H 2 A. Z promotes the formation of more open chromatin and facilitates repair from Volle and Dalal, Curr. Opin. Genet. Dev. 25, 8 (2014)

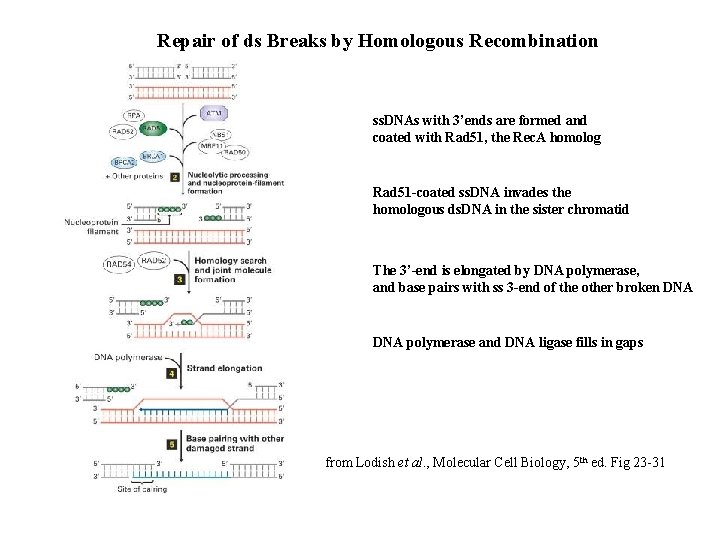
Repair of ds Breaks by Homologous Recombination ss. DNAs with 3’ends are formed and coated with Rad 51, the Rec. A homolog Rad 51 -coated ss. DNA invades the homologous ds. DNA in the sister chromatid The 3’-end is elongated by DNA polymerase, and base pairs with ss 3 -end of the other broken DNA polymerase and DNA ligase fills in gaps from Lodish et al. , Molecular Cell Biology, 5 th ed. Fig 23 -31

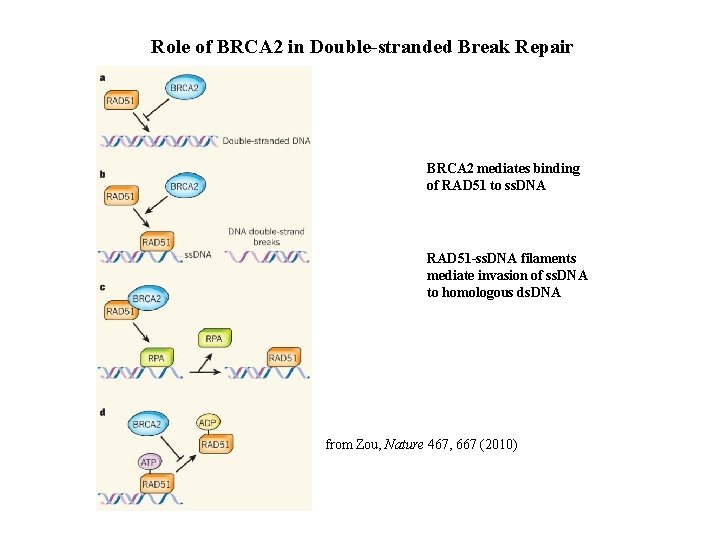
Role of BRCA 2 in Double-stranded Break Repair BRCA 2 mediates binding of RAD 51 to ss. DNA RAD 51 -ss. DNA filaments mediate invasion of ss. DNA to homologous ds. DNA from Zou, Nature 467, 667 (2010)

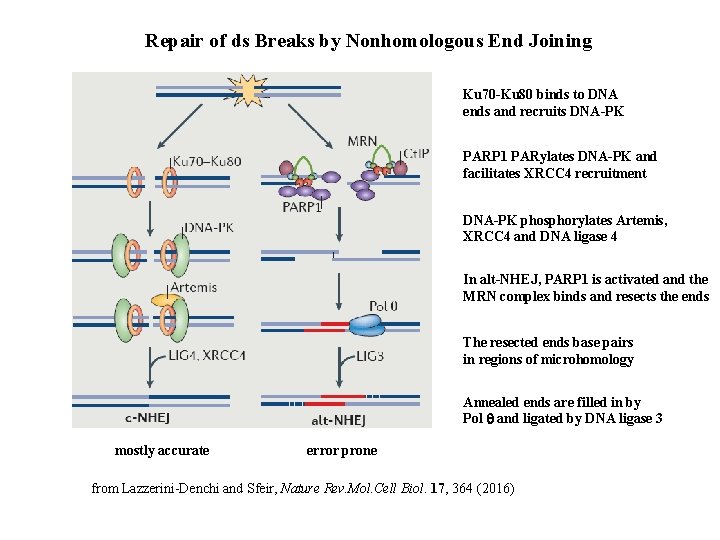
Repair of ds Breaks by Nonhomologous End Joining Ku 70 -Ku 80 binds to DNA ends and recruits DNA-PK PARP 1 PARylates DNA-PK and facilitates XRCC 4 recruitment DNA-PK phosphorylates Artemis, XRCC 4 and DNA ligase 4 In alt-NHEJ, PARP 1 is activated and the MRN complex binds and resects the ends The resected ends base pairs in regions of microhomology Annealed ends are filled in by Pol q and ligated by DNA ligase 3 mostly accurate error prone from Lazzerini-Denchi and Sfeir, Nature Rev. Mol. Cell Biol. 17, 364 (2016)

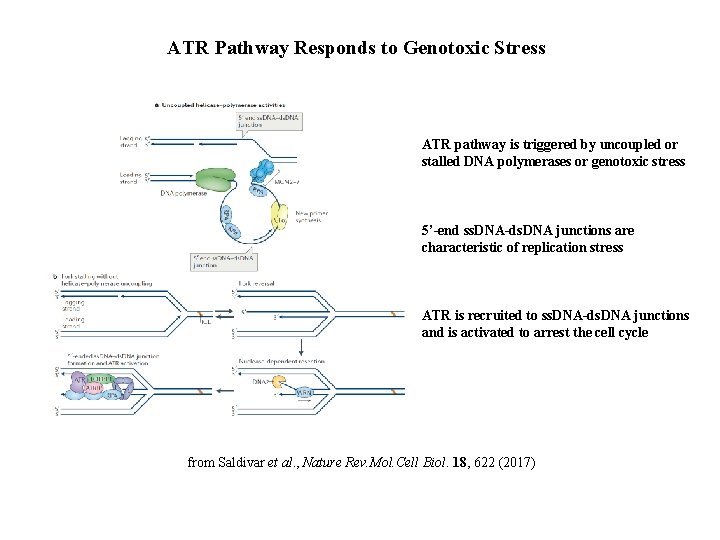
ATR Pathway Responds to Genotoxic Stress ATR pathway is triggered by uncoupled or stalled DNA polymerases or genotoxic stress 5’-end ss. DNA-ds. DNA junctions are characteristic of replication stress ATR is recruited to ss. DNA-ds. DNA junctions and is activated to arrest the cell cycle from Saldivar et al. , Nature Rev. Mol. Cell Biol. 18, 622 (2017)

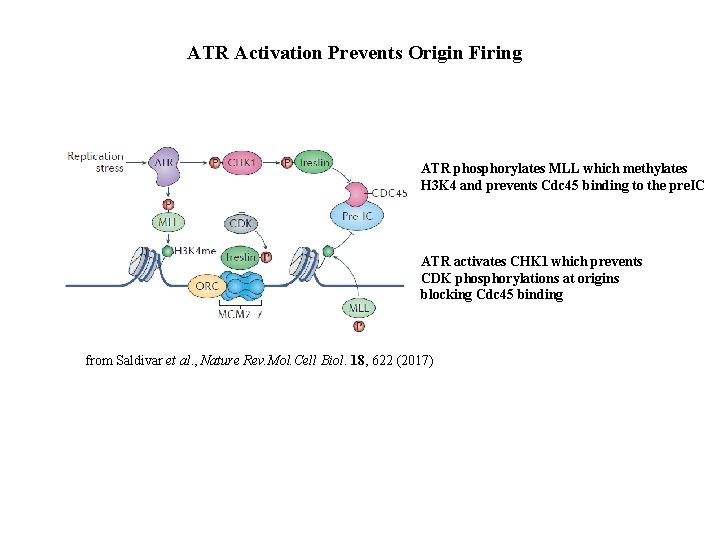
ATR Activation Prevents Origin Firing ATR phosphorylates MLL which methylates H 3 K 4 and prevents Cdc 45 binding to the pre. IC ATR activates CHK 1 which prevents CDK phosphorylations at origins blocking Cdc 45 binding from Saldivar et al. , Nature Rev. Mol. Cell Biol. 18, 622 (2017)

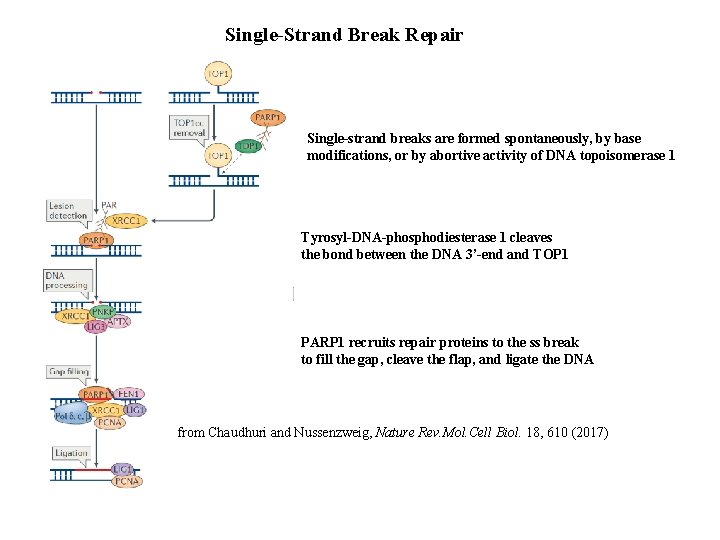
Single-Strand Break Repair Single-strand breaks are formed spontaneously, by base modifications, or by abortive activity of DNA topoisomerase 1 Tyrosyl-DNA-phosphodiesterase 1 cleaves the bond between the DNA 3’-end and TOP 1 PARP 1 recruits repair proteins to the ss break to fill the gap, cleave the flap, and ligate the DNA from Chaudhuri and Nussenzweig, Nature Rev. Mol. Cell Biol. 18, 610 (2017)

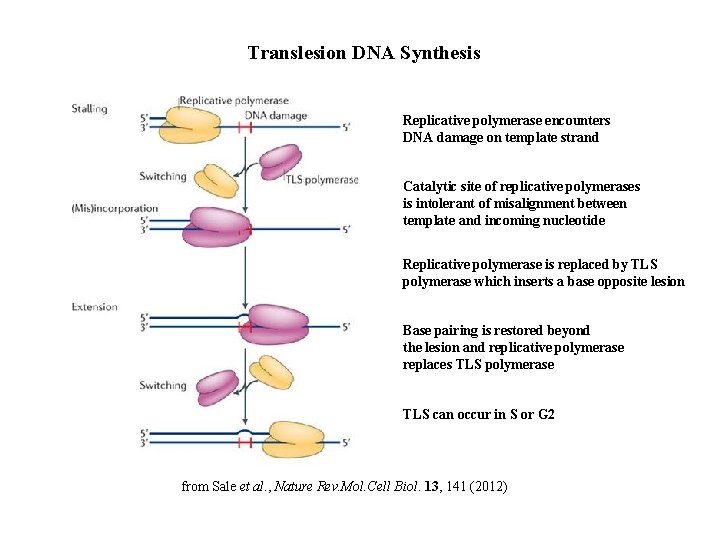
Translesion DNA Synthesis Replicative polymerase encounters DNA damage on template strand Catalytic site of replicative polymerases is intolerant of misalignment between template and incoming nucleotide Replicative polymerase is replaced by TLS polymerase which inserts a base opposite lesion Base pairing is restored beyond the lesion and replicative polymerase replaces TLS polymerase TLS can occur in S or G 2 from Sale et al. , Nature Rev. Mol. Cell Biol. 13, 141 (2012)

There are Multiple TLS Polymerases TLS polymerases are recruited by interactions with the sliding clamp There are multiple TLS polymerases have low processivity and low fidelity, and lack 3’-5’ exonucleases TLS polymerases are selective for certain lesions from Sale et al. , Nature Rev. Mol. Cell Biol. 13, 141 (2012) Most mutations caused by DNA lesions are caused by TLS polymerases

TLS Polymerases Can Be Accurate or Error-prone Pol k bypasses an abasic site and often causes a -1 frameshift Pol h bypasses a thymine dimer and inserts AA Pol i is accurate with d. A template and error-prone with d. T template Replicative polymerases insert d. C or d. A opposite 8 -oxo-G, Pol i inserts d. C The likelihood that TLS polymerases are error-prone depends on the nature of the lesion and the TLS polymerase that is utilized

Somatic Hypermutation of Ig Genes Depends on TLS Polymerases AID deaminates d. C to d. U Uracil DNA glycosylase forms an abasic site, and REV 1 incorporates d. C opposite the site MMR proteins lead to the formation of a ss gap, PCNA is ubiquitylated, and Pol h is recruited, generating mutations at A-T from Sale et al. , Nature Rev. Mol. Cell Biol. 13, 141 (2012)
 Coding dna and non coding dna
Coding dna and non coding dna Enzyme involved in dna replication
Enzyme involved in dna replication Replication
Replication Dna and genes chapter 11
Dna and genes chapter 11 Bioflix activity dna replication nucleotide pairing
Bioflix activity dna replication nucleotide pairing Chart
Chart Bilirubin bruise
Bilirubin bruise Inhibitor of oxidative phosphorylation
Inhibitor of oxidative phosphorylation Urocanate reductase
Urocanate reductase Oxidative phosphorylation energy yield
Oxidative phosphorylation energy yield Corrosion principle
Corrosion principle Uncouplers of oxidative phosphorylation
Uncouplers of oxidative phosphorylation P 450
P 450 Oxidation desizing agent
Oxidation desizing agent Uncouplers of oxidative phosphorylation
Uncouplers of oxidative phosphorylation Phosphorylation definition
Phosphorylation definition Deamination of glutamine
Deamination of glutamine Uncouplers of oxidative phosphorylation
Uncouplers of oxidative phosphorylation Pentose phosphate pathway
Pentose phosphate pathway Amino acids in citric acid cycle
Amino acids in citric acid cycle Pentose phosphate pathway enzymes
Pentose phosphate pathway enzymes Amino urea
Amino urea Inhibitors of oxidative phosphorylation
Inhibitors of oxidative phosphorylation Concept map about oxidative phosphorylation
Concept map about oxidative phosphorylation Glutamate oxidative deamination
Glutamate oxidative deamination Fructose breakdown pathway
Fructose breakdown pathway Oxidative phosphorylation
Oxidative phosphorylation Substrate level phosphorylation
Substrate level phosphorylation Oxidative deamination of amino acids
Oxidative deamination of amino acids Oxidative addition of h2
Oxidative addition of h2 Transamination and oxidative deamination
Transamination and oxidative deamination Decarboxylation in krebs cycle
Decarboxylation in krebs cycle Chapter 9: cellular respiration: harvesting chemical energy
Chapter 9: cellular respiration: harvesting chemical energy Uncouple oxidative phosphorylation
Uncouple oxidative phosphorylation Define oxidative phosphorylation
Define oxidative phosphorylation Energetics of oxidative phosphorylation
Energetics of oxidative phosphorylation Substrate level phosphorylation vs oxidative
Substrate level phosphorylation vs oxidative Contract ld
Contract ld Crashworthiness course
Crashworthiness course Flood damage restoration the basin
Flood damage restoration the basin Sarasota storm damage
Sarasota storm damage Tigh
Tigh Stages of damage control surgery
Stages of damage control surgery Trailer equipment damage reduction
Trailer equipment damage reduction