DNA Repair DNA repair refers to a collection















- Slides: 30

DNA Repair

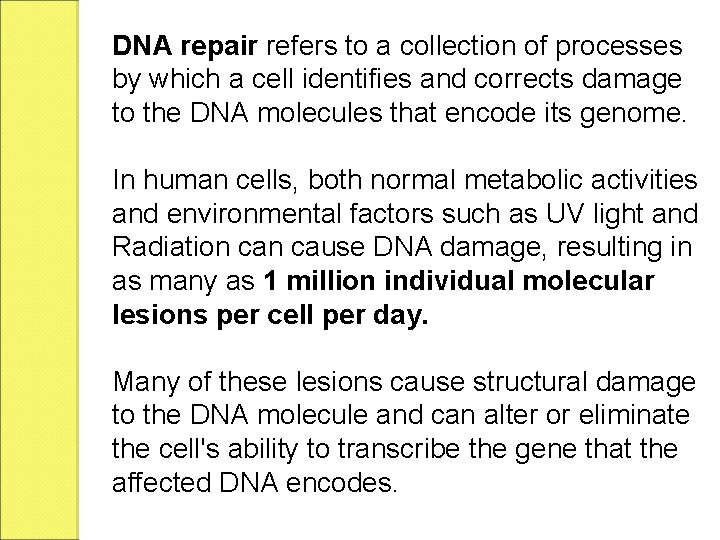
DNA repair refers to a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as UV light and Radiation cause DNA damage, resulting in as many as 1 million individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes.

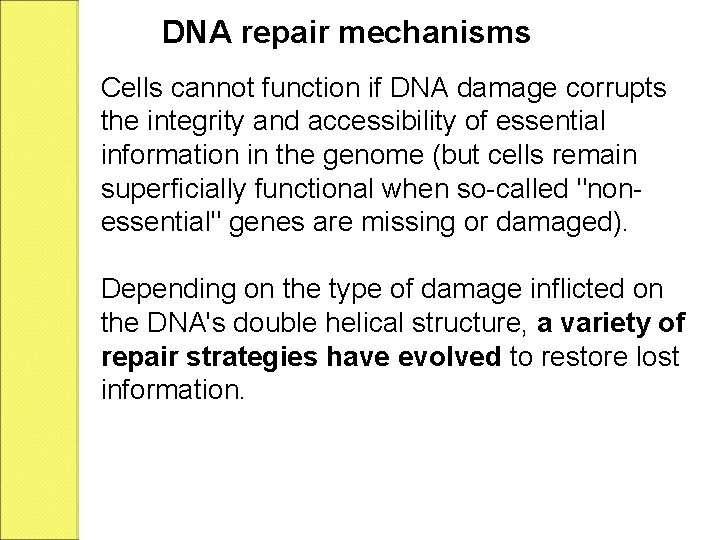
DNA repair mechanisms Cells cannot function if DNA damage corrupts the integrity and accessibility of essential information in the genome (but cells remain superficially functional when so-called "nonessential" genes are missing or damaged). Depending on the type of damage inflicted on the DNA's double helical structure, a variety of repair strategies have evolved to restore lost information.


Direct reversal Cells are known to eliminate damage to their DNA by chemically reversing it. These mechanisms do not require a template, since the types of damage they counteract can only occur in one of the four bases. Such direct reversal mechanisms are specific to the type of damage incurred and do not involve breakage of the phosphodiester backbone.

An example: methylation of guanine bases, is directly reversed by the protein methyl guanine methyl transferase (MGMT), the bacterial equivalent of which is called as ogt. This is an expensive process because each MGMT molecule can only be used once; that is, the reaction is stoichiometric rather than catalytic.

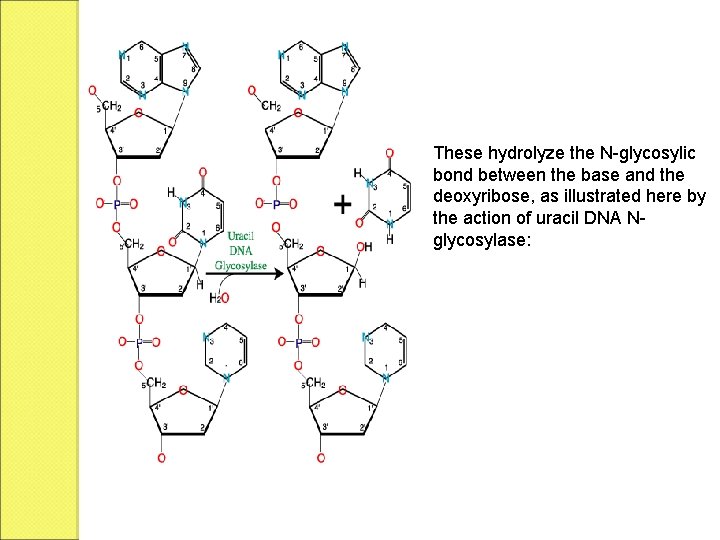
1. Base excision repair (BER), which repairs damage to a single base caused by oxidation, alkylation, hydrolysis, or deamination. The damaged base is removed by a DNA glycosylase, resynthesized by a DNA polymerase, and a DNA ligase performs the final nick-sealing step.

These hydrolyze the N-glycosylic bond between the base and the deoxyribose, as illustrated here by the action of uracil DNA Nglycosylase:

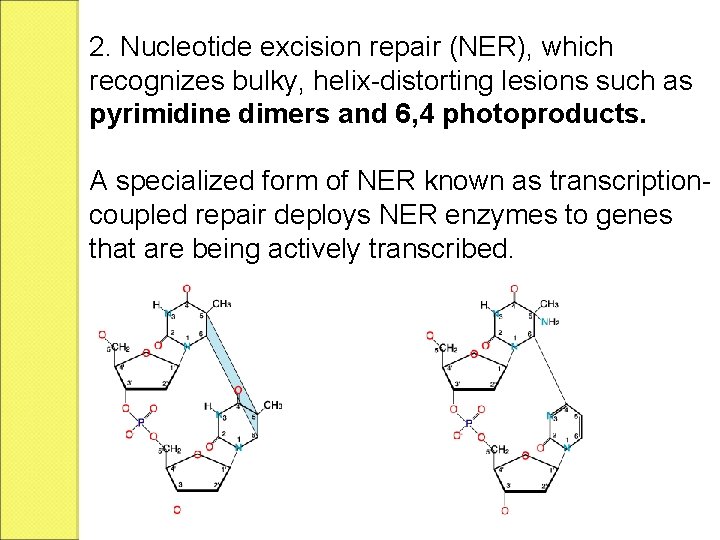
2. Nucleotide excision repair (NER), which recognizes bulky, helix-distorting lesions such as pyrimidine dimers and 6, 4 photoproducts. A specialized form of NER known as transcriptioncoupled repair deploys NER enzymes to genes that are being actively transcribed.

NER involves the following steps: Damage recognition Binding of a multi-protein complex at the damaged site Double incision of the damaged strand several nucleotides away from the damaged site, on both the 5' and 3' sides Removal of the damage-containing oligonucleotide from between the two nicks Filling in of the resulting gap by a DNA polymerase Ligation



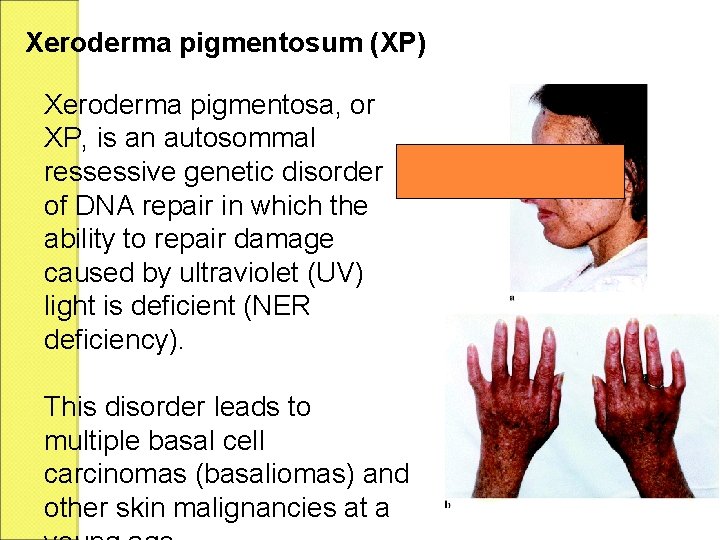
Xeroderma pigmentosum (XP) Xeroderma pigmentosa, or XP, is an autosommal ressessive genetic disorder of DNA repair in which the ability to repair damage caused by ultraviolet (UV) light is deficient (NER deficiency). This disorder leads to multiple basal cell carcinomas (basaliomas) and other skin malignancies at a

In severe cases, it is necessary to avoid sunlight completely. The two most common causes of death for XP victims are metastatic malignant melanoma and squamous cell carcinoma.

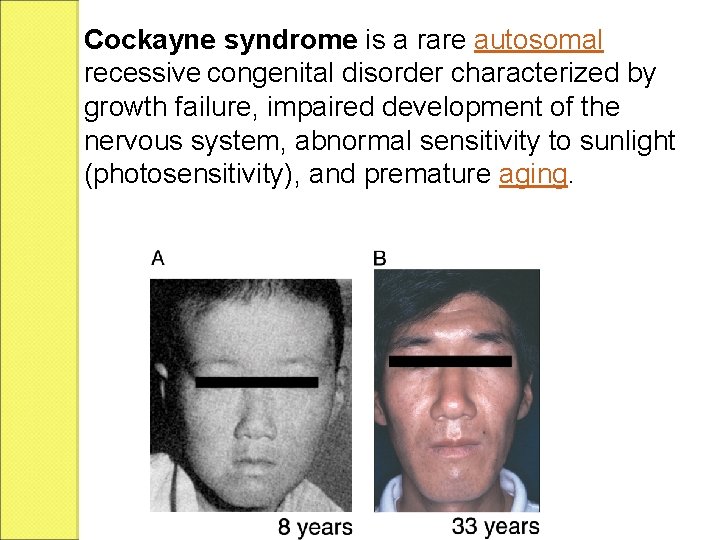
Cockayne syndrome is a rare autosomal recessive congenital disorder characterized by growth failure, impaired development of the nervous system, abnormal sensitivity to sunlight (photosensitivity), and premature aging.

3. Mismatch repair (MMR), which corrects errors of DNA replication and recombination that result in mispaired (but undamaged) nucleotides.


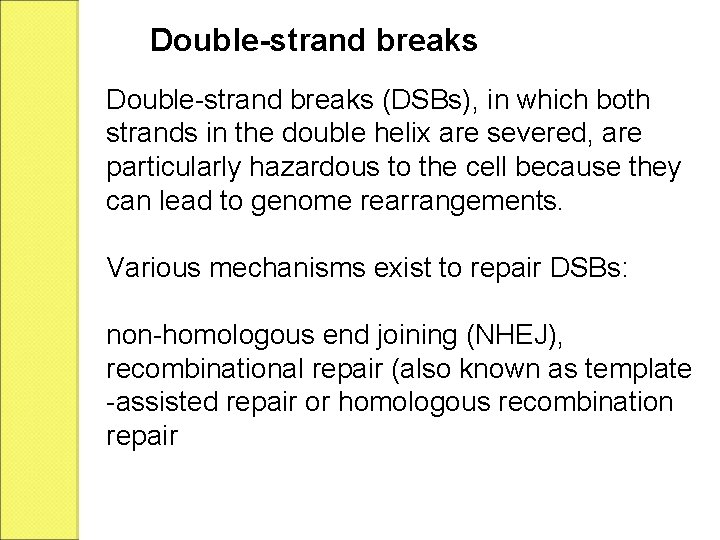
Double-strand breaks (DSBs), in which both strands in the double helix are severed, are particularly hazardous to the cell because they can lead to genome rearrangements. Various mechanisms exist to repair DSBs: non-homologous end joining (NHEJ), recombinational repair (also known as template -assisted repair or homologous recombination repair


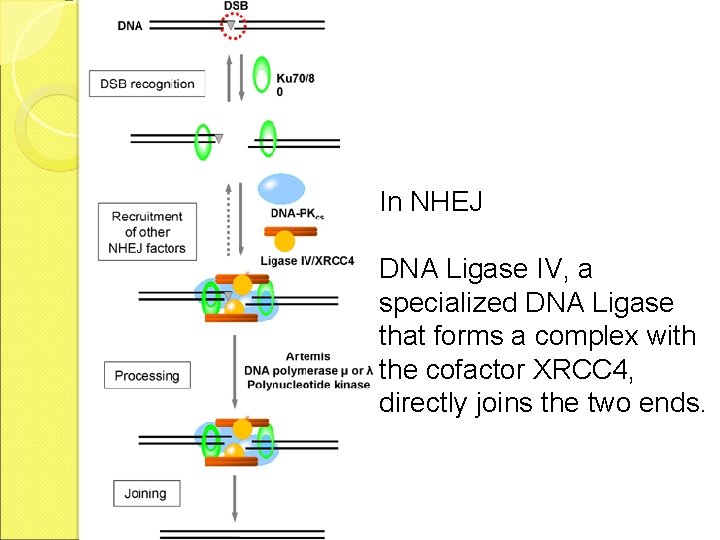
In NHEJ DNA Ligase IV, a specialized DNA Ligase that forms a complex with the cofactor XRCC 4, directly joins the two ends.

DNA ligase, shown above repairing chromosomal damage, is an enzyme that joins broken nucleotides together by catalyzing the formation of an internucleotide ester bond between the phosphate backbone and the deoxyribose nucleotides.

Recombinational Repair Recombinational repair requires the presence of an identical or nearly identical sequence to be used as a template for repair of the break. The enzymatic machinery responsible for this repair process is nearly identical to the machinery responsible for chromosomal crossover during meiosis. This pathway allows a damaged chromosome to be repaired using a sister chromatid (available in G 2 after DNA replication) or a homologous chromosome as



Translesion synthesis is a DNA damage tolerance process that allows the DNA replication machinery to replicate past DNA lesions such as thymine dimers or AP sites. It involves switching out regular DNA polymerases for specialized translesion polymerases (e. g. DNA polymerase V), often with larger active sites that can facilitate the insertion of bases opposite damaged nucleotides.

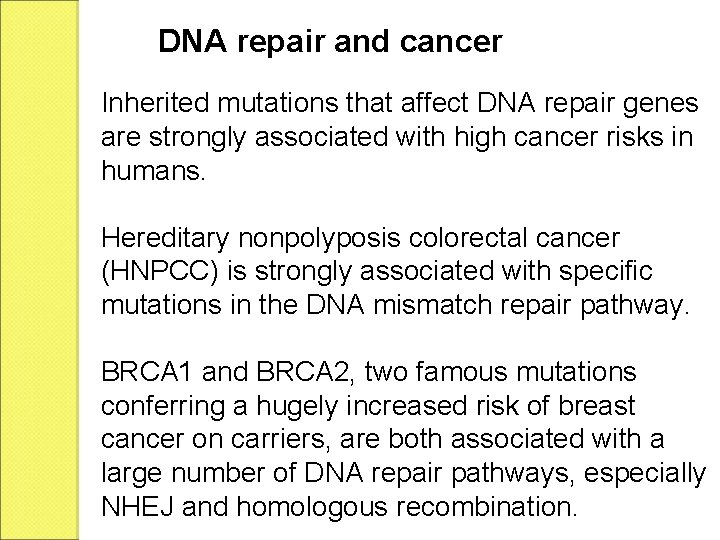
The polymerase switching is thought to be mediated by, among other factors, the post-translational modification of the replication processivity factor PCNA. Translesion synthesis polymerases often have low fidelity (high propensity to insert wrong bases) relative to regular polymerases. PCNA Proliferating Cell Nuclear Antigen

DNA repair and cancer Inherited mutations that affect DNA repair genes are strongly associated with high cancer risks in humans. Hereditary nonpolyposis colorectal cancer (HNPCC) is strongly associated with specific mutations in the DNA mismatch repair pathway. BRCA 1 and BRCA 2, two famous mutations conferring a hugely increased risk of breast cancer on carriers, are both associated with a large number of DNA repair pathways, especially NHEJ and homologous recombination.

Modern cancer treatments attempt to localize the DNA damage to cells and tissues only associated with cancer, either by physical means (concentrating therapeutic agent in the region of the tumor) or by biochemical means (exploiting a feature unique to cancer cells in the body).

Cancer Chemotherapy The hallmark of all cancers is continuous cell division. Each division requires both the replication of the cell's DNA (in S phase) and transcription and translation of many genes needed for continued growth. So, any chemical that damages DNA has the potential to inhibit the spread of a cancer. Many (but not all) drugs used for cancer therapy do their work by damaging DNA.

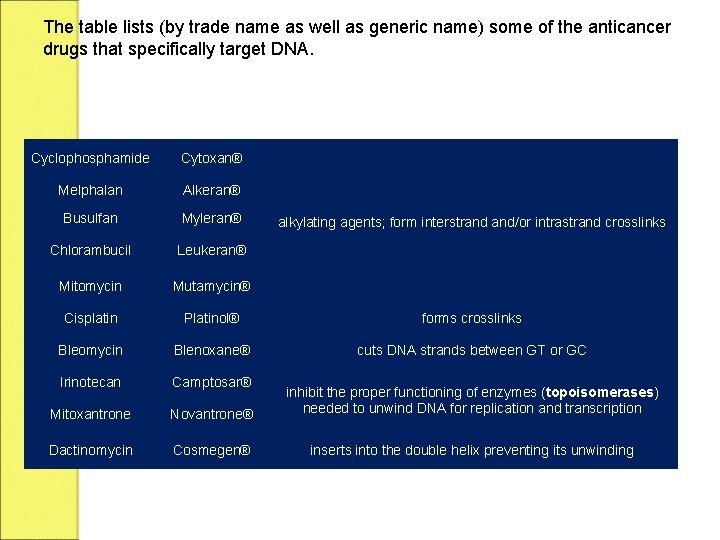
The table lists (by trade name as well as generic name) some of the anticancer drugs that specifically target DNA. Cyclophosphamide Cytoxan® Melphalan Alkeran® Busulfan Myleran® Chlorambucil Leukeran® Mitomycin Mutamycin® Cisplatin Platinol® forms crosslinks Bleomycin Blenoxane® cuts DNA strands between GT or GC Irinotecan Camptosar® Mitoxantrone Novantrone® inhibit the proper functioning of enzymes (topoisomerases) needed to unwind DNA for replication and transcription Dactinomycin Cosmegen® inserts into the double helix preventing its unwinding alkylating agents; form interstrand and/or intrastrand crosslinks
 Rec a
Rec a Base excision repair vs mismatch repair
Base excision repair vs mismatch repair Landsat collection 1 vs collection 2
Landsat collection 1 vs collection 2 Types of documentary collection
Types of documentary collection Dna polymerase proofreading
Dna polymerase proofreading 반보존적 복제
반보존적 복제 Coding dna and non coding dna
Coding dna and non coding dna The principal enzyme involved in dna replication is
The principal enzyme involved in dna replication is Function of dna polymerase 3
Function of dna polymerase 3 Dna rna protein synthesis homework #2 dna replication
Dna rna protein synthesis homework #2 dna replication Bioflix activity dna replication lagging strand synthesis
Bioflix activity dna replication lagging strand synthesis Electronic suspension system
Electronic suspension system Phrase tree diagrams
Phrase tree diagrams Level o dfd
Level o dfd Laceration repair irving
Laceration repair irving Medial meniscus root repair
Medial meniscus root repair No verb
No verb Penapis
Penapis Az vm repair create
Az vm repair create Facial nerve repair
Facial nerve repair Prostrct repair
Prostrct repair Sefpro repair services
Sefpro repair services Tissue repair
Tissue repair Draw a binary search tree for the sentence
Draw a binary search tree for the sentence Cpvc ductwork
Cpvc ductwork Chapter 44 automotive wiring and wire repair
Chapter 44 automotive wiring and wire repair Slab leak repair beaver creek
Slab leak repair beaver creek Chapter 10 lesson 2 nutrients
Chapter 10 lesson 2 nutrients Isri 6860 repair manual
Isri 6860 repair manual Repair and restoration theory of sleep
Repair and restoration theory of sleep Crack comparator card
Crack comparator card