North American Cooperation in Genomic Prediction Paul Van







- Slides: 34

North American Cooperation in Genomic Prediction Paul Van. Raden 1, George Wiggans 1, Curt Van Tassell 2, Tad Sonstegard 2, Jeff O’Connell 1, Bob Schnabel 3, Jerry Taylor 3, and Flavio Schenkel 4, 1 Animal Improvement Programs Lab and 2 Bovine Functional Genomics Lab, USDA Agricultural Research Service, Beltsville, MD, USA, 3 U. Missouri, Columbia, 4 U. Guelph, ON, Canada Paul. Van. Raden@ars. usda. gov 2008 2007

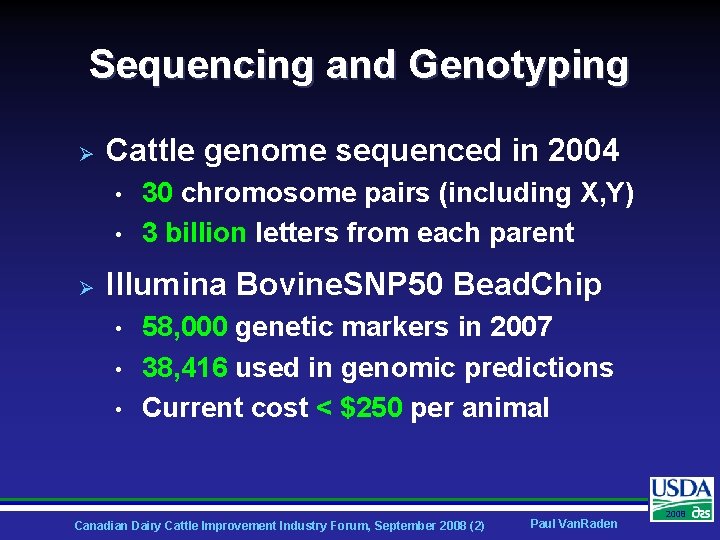
Sequencing and Genotyping Ø Cattle genome sequenced in 2004 • • Ø 30 chromosome pairs (including X, Y) 3 billion letters from each parent Illumina Bovine. SNP 50 Bead. Chip • • • 58, 000 genetic markers in 2007 38, 416 used in genomic predictions Current cost < $250 per animal Canadian Dairy Cattle Improvement Industry Forum, September 2008 (2) Paul Van. Raden 2008

History of Genomic Cooperation Ø 1992 USDA request for CAN, USA AI companies to join Dairy Bull DNA Repository maintained at U. Illinois • Ø CIAQ (Brian Van Doormaal) provided 1110 straws from 110 bulls 1999 Cooperative Dairy DNA Repository maintained at Beltsville, MD • Semex and Alta contributed semen for all progeny tested bulls Canadian Dairy Cattle Improvement Industry Forum, September 2008 (3) Paul Van. Raden 2008

CDDR Contributors Ø National Association of Animal Breeders (NAAB, Columbia, MO) • ABS Global (De. Forest, WI) Accelerated Genetics (Baraboo, WI) Alta (Balzac, AB) Genex (Shawano, WI) New Generation Genetics (Fort Atkinson, WI) • Select Sires (Plain City, OH) • Semex Alliance (Guelph, ON) • Taurus-Service (Mehoopany, PA) • • Canadian Dairy Cattle Improvement Industry Forum, September 2008 (4) Paul Van. Raden 2008

Genotype Data for Elevation Chromosome 1 10001112200200121110111121111001121100020122002220111 1202101200211122110021112001111001011011010220011002201101 12002011010202221211221020100111000112202212221120120 201002022020000211000112020112211102201111000021220200 0221012020002211220111012100111211102112110020102100022000 2201000201100002202211210112111012222001211212220020020202012221100222222200221211112100211112001101120 0202220001112011010211121211102022100211201211001111102111 2110211122000101101110202200221110102011121111011202102102 121101102212200121101202201100222002110001110021 1021101110002220020221212110002220102002222121221121112002 0110202001222222112212021211210110012110110200220002001002 000111101100121102121211120101012120221010101111102112 211111121211121011012001111102111101111122012121101022 202021211222120222002121210201100111222121101 Canadian Dairy Cattle Improvement Industry Forum, September 2008 (5) Paul Van. Raden 2008

Genotype Data from Inbred Bull Chromosome 24 of Megastar 1021222101021021011102110112112110022020002020220 000022002022220220200002002022222200002022220000022020000200200000022220000002022002000222020222220002 2022222000020022020222020002200002202220000002200 2020002222002002020222222220200020220220222202022000222022002220000022020000200200200222220 0022220202002220200000022222020200002002002222000 2022022220002222022002222020200022022022220022000200 220200000220222000022000222202002222000220020020202 202000222002220220220000022022002002002022000222202 200222002022020022220000020220002020200022000002 2022200202220200022002002000200220222220022022000020000202202002000022200200222000022 02200200220220202020002220200022020020220220000 20202000020202000222222000200220222200000202200202 02202202020002002202200 Canadian Dairy Cattle Improvement Industry Forum, September 2008 (6) Paul Van. Raden 2008

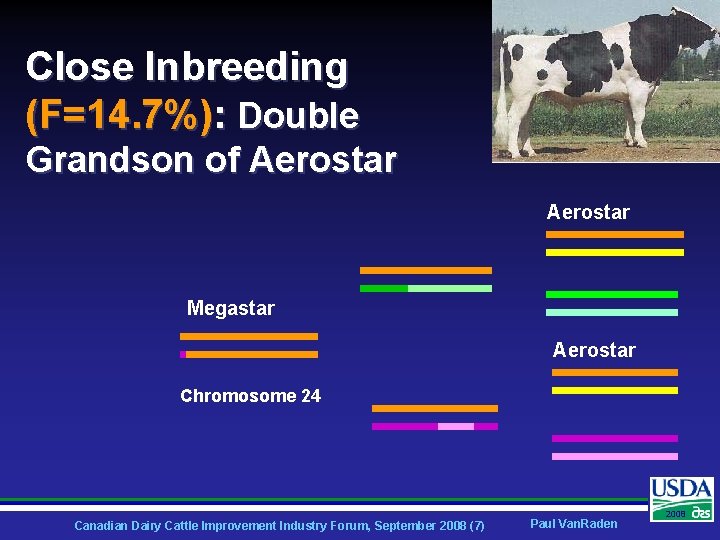
Close Inbreeding (F=14. 7%): Double Grandson of Aerostar Megastar Aerostar Chromosome 24 Canadian Dairy Cattle Improvement Industry Forum, September 2008 (7) Paul Van. Raden 2008

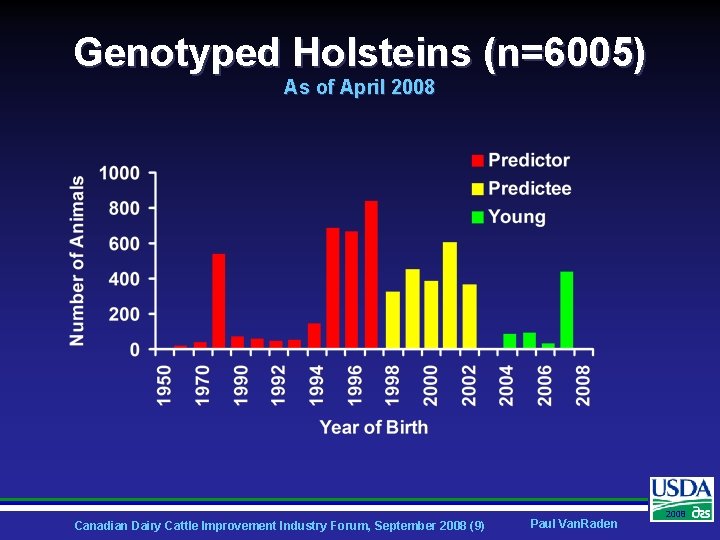
Holstein Experimental Design Ø Compute genomic evaluations and parent averages from 2003 data • Ø Compare ability to predict daughter deviations in 2008 data • Ø 3576 older Holstein bulls born 1952 -1998 1759 younger bulls born 1999 -2002 Test results for 27 traits: 5 yield, 5 health, 16 conformation, and Net Merit Canadian Dairy Cattle Improvement Industry Forum, September 2008 (8) Paul Van. Raden 2008

Genotyped Holsteins (n=6005) As of April 2008 Canadian Dairy Cattle Improvement Industry Forum, September 2008 (9) Paul Van. Raden 2008

Jersey and Brown Swiss Ø Jersey experimental design • • Ø Data from 579 bulls born before 1999 and 204 cows with data before 2003 Predict 352 bulls born during or after 1999 Brown Swiss experimental design • • Data from 225 bulls born before 1999 Predict 118 bulls born during or after 1999 Canadian Dairy Cattle Improvement Industry Forum, September 2008 (10) Paul Van. Raden 2008

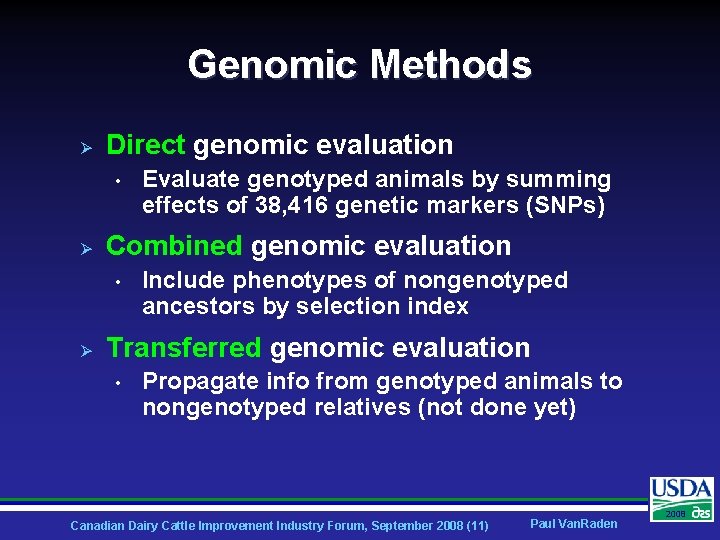
Genomic Methods Ø Direct genomic evaluation • Ø Combined genomic evaluation • Ø Evaluate genotyped animals by summing effects of 38, 416 genetic markers (SNPs) Include phenotypes of nongenotyped ancestors by selection index Transferred genomic evaluation • Propagate info from genotyped animals to nongenotyped relatives (not done yet) Canadian Dairy Cattle Improvement Industry Forum, September 2008 (11) Paul Van. Raden 2008

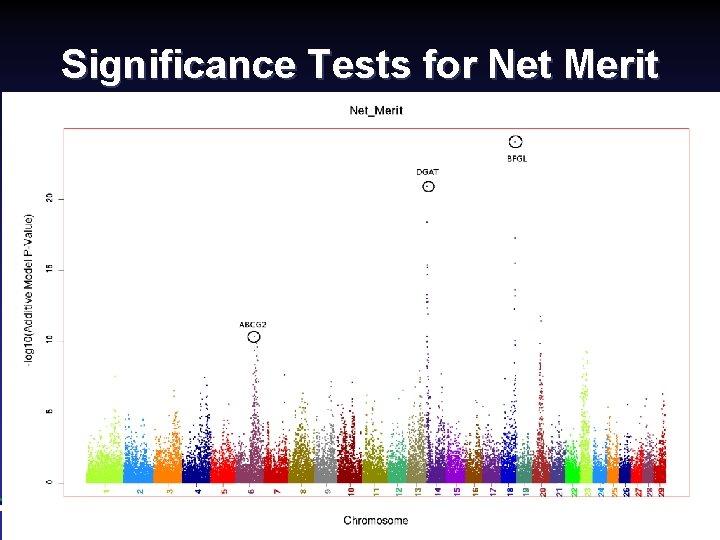
Significance Tests for Net Merit Canadian Dairy Cattle Improvement Industry Forum, September 2008 (12) Paul Van. Raden 2008

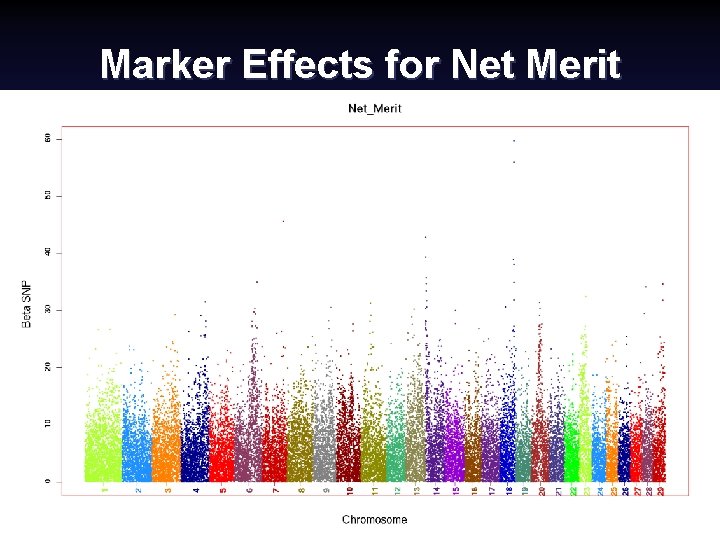
Marker Effects for Net Merit Canadian Dairy Cattle Improvement Industry Forum, September 2008 (13) Paul Van. Raden 2008

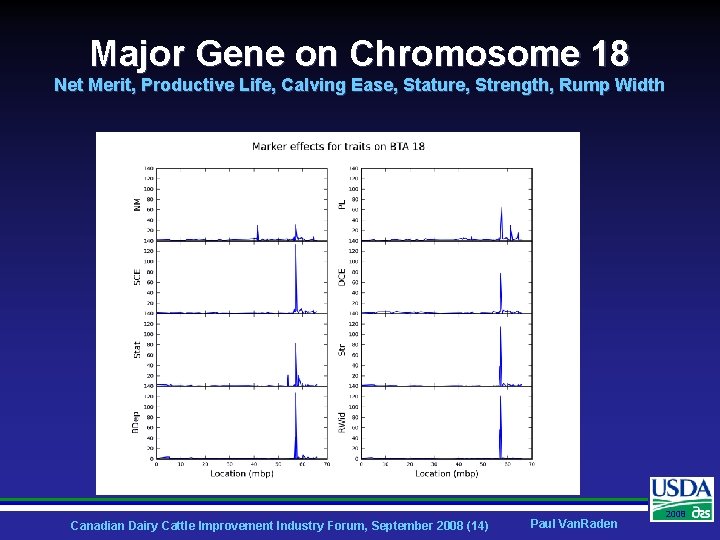
Major Gene on Chromosome 18 Net Merit, Productive Life, Calving Ease, Stature, Strength, Rump Width Canadian Dairy Cattle Improvement Industry Forum, September 2008 (14) Paul Van. Raden 2008

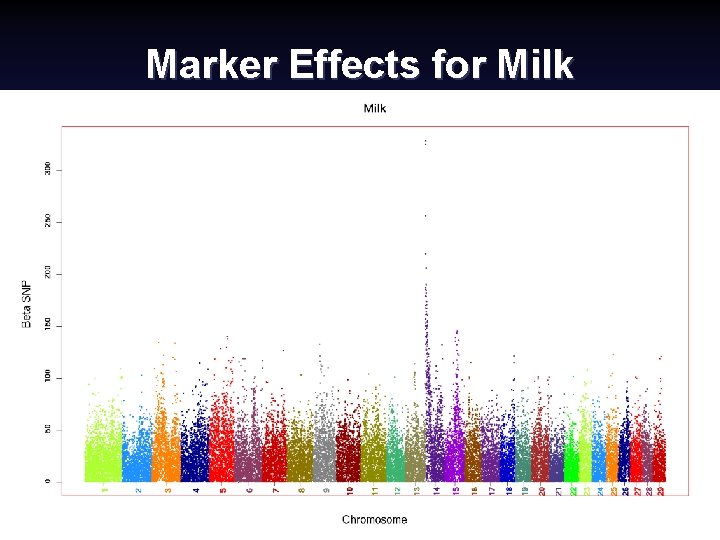
Marker Effects for Milk Canadian Dairy Cattle Improvement Industry Forum, September 2008 (15) Paul Van. Raden 2008

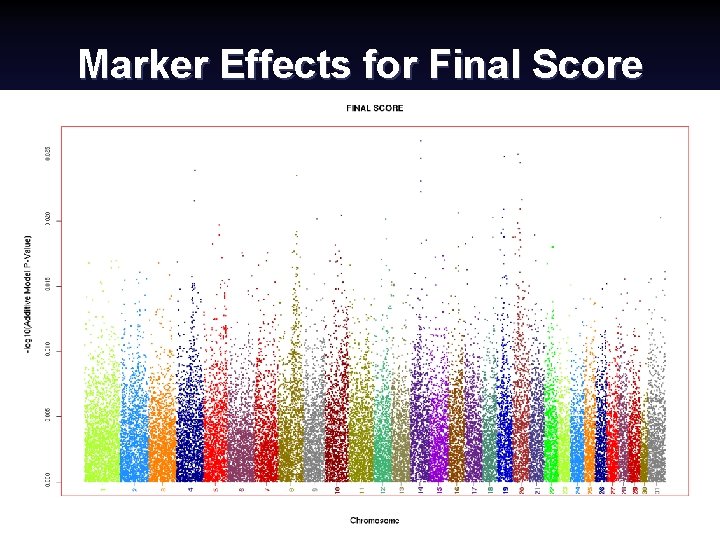
Marker Effects for Final Score Canadian Dairy Cattle Improvement Industry Forum, September 2008 (16) Paul Van. Raden 2008

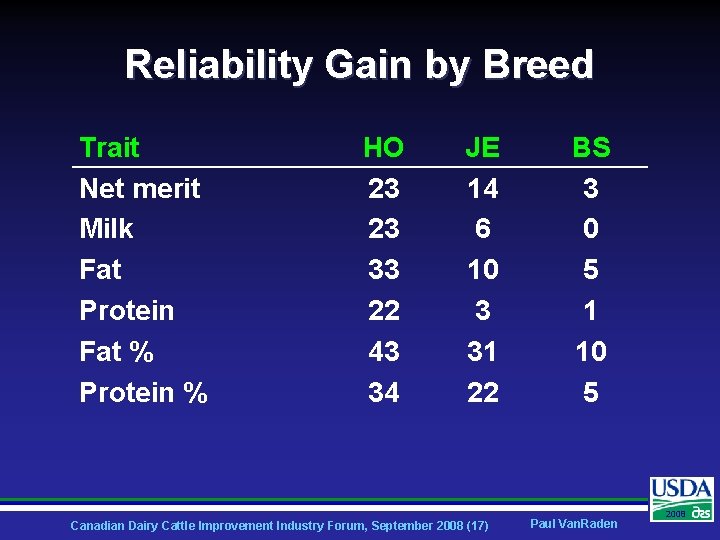
Reliability Gain by Breed Trait Net merit Milk Fat Protein Fat % Protein % HO 23 23 33 22 43 34 JE 14 6 10 3 31 22 Canadian Dairy Cattle Improvement Industry Forum, September 2008 (17) BS 3 0 5 1 10 5 Paul Van. Raden 2008

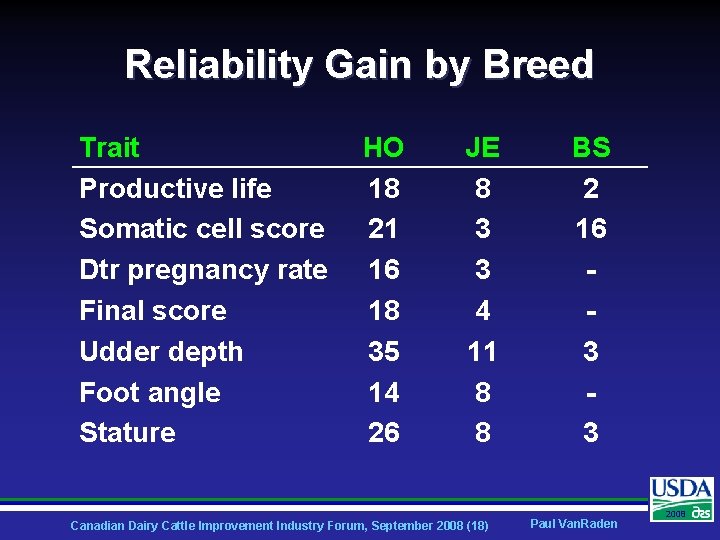
Reliability Gain by Breed Trait Productive life Somatic cell score Dtr pregnancy rate Final score Udder depth Foot angle Stature HO 18 21 16 18 35 14 26 JE 8 3 3 4 11 8 8 Canadian Dairy Cattle Improvement Industry Forum, September 2008 (18) BS 2 16 3 3 Paul Van. Raden 2008

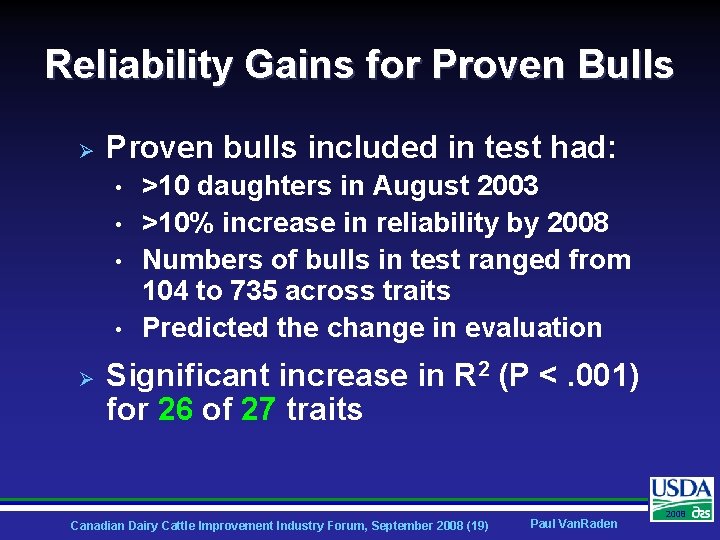
Reliability Gains for Proven Bulls Ø Proven bulls included in test had: • • Ø >10 daughters in August 2003 >10% increase in reliability by 2008 Numbers of bulls in test ranged from 104 to 735 across traits Predicted the change in evaluation Significant increase in R 2 (P <. 001) for 26 of 27 traits Canadian Dairy Cattle Improvement Industry Forum, September 2008 (19) Paul Van. Raden 2008

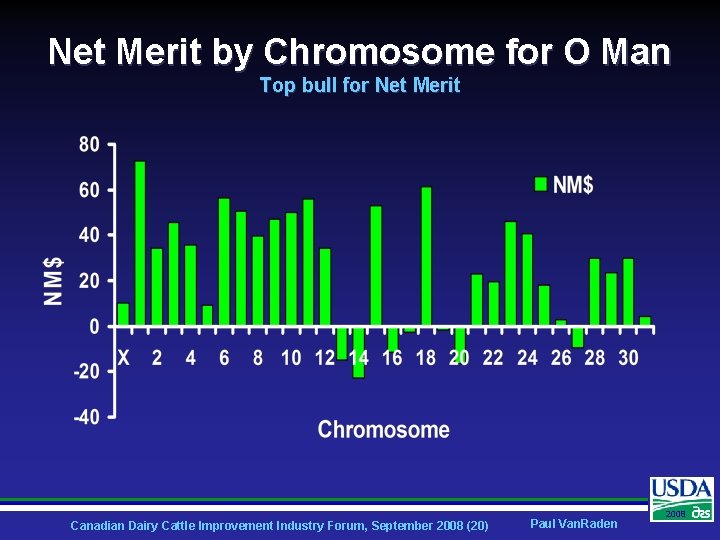
Net Merit by Chromosome for O Man Top bull for Net Merit Canadian Dairy Cattle Improvement Industry Forum, September 2008 (20) Paul Van. Raden 2008

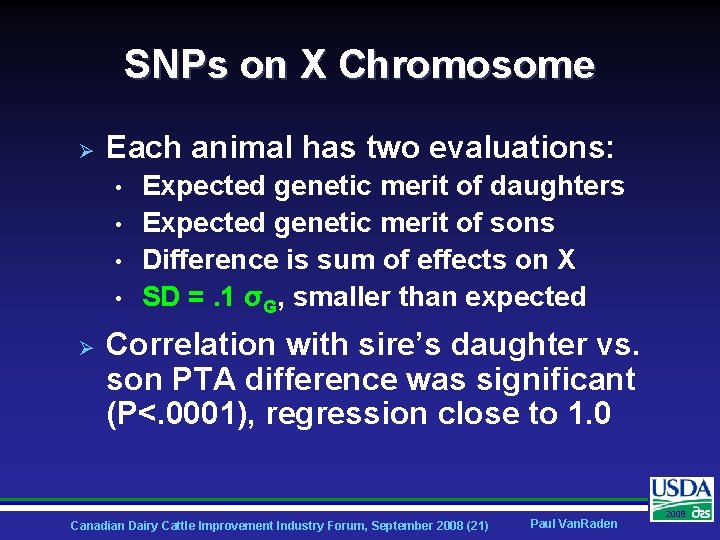
SNPs on X Chromosome Ø Each animal has two evaluations: • • Ø Expected genetic merit of daughters Expected genetic merit of sons Difference is sum of effects on X SD =. 1 σG, smaller than expected Correlation with sire’s daughter vs. son PTA difference was significant (P<. 0001), regression close to 1. 0 Canadian Dairy Cattle Improvement Industry Forum, September 2008 (21) Paul Van. Raden 2008

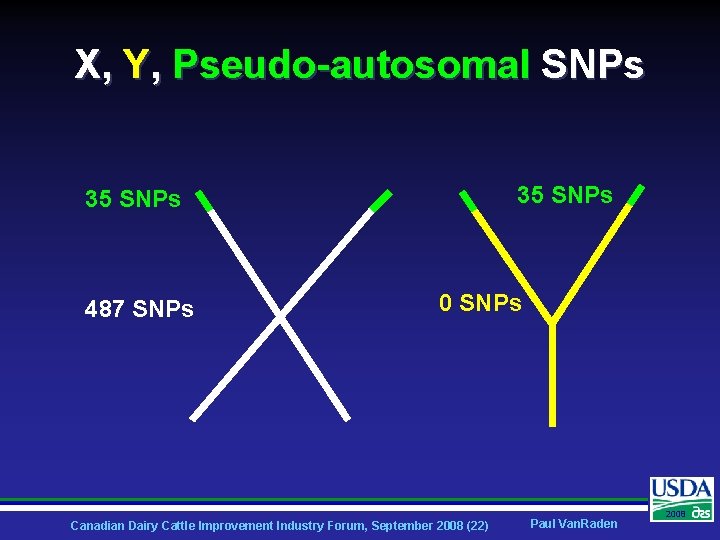
X, Y, Pseudo-autosomal SNPs 35 SNPs 487 SNPs 0 SNPs Canadian Dairy Cattle Improvement Industry Forum, September 2008 (22) Paul Van. Raden 2008

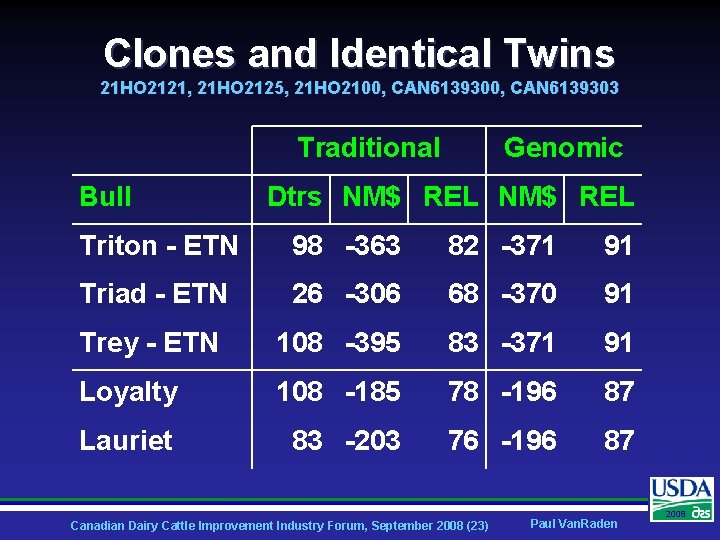
Clones and Identical Twins 21 HO 2121, 21 HO 2125, 21 HO 2100, CAN 6139303 Traditional Bull Genomic Dtrs NM$ REL Triton - ETN 98 -363 82 -371 91 Triad - ETN 26 -306 68 -370 91 Trey - ETN 108 -395 83 -371 91 Loyalty 108 -185 78 -196 87 Lauriet 83 -203 76 -196 87 Canadian Dairy Cattle Improvement Industry Forum, September 2008 (23) Paul Van. Raden 2008

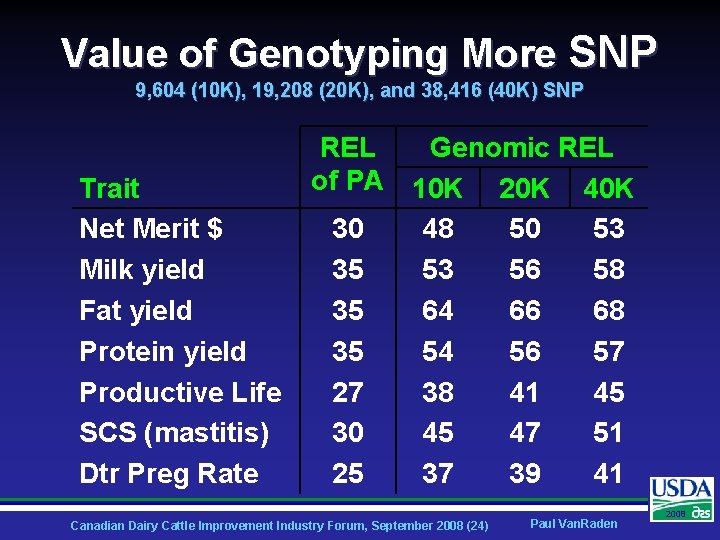
Value of Genotyping More SNP 9, 604 (10 K), 19, 208 (20 K), and 38, 416 (40 K) SNP Trait Net Merit $ Milk yield Fat yield Protein yield Productive Life SCS (mastitis) Dtr Preg Rate REL Genomic REL of PA 10 K 20 K 40 K 30 48 50 53 35 53 56 58 35 64 66 68 35 54 56 57 27 38 41 45 30 45 47 51 25 37 39 41 Canadian Dairy Cattle Improvement Industry Forum, September 2008 (24) Paul Van. Raden 2008

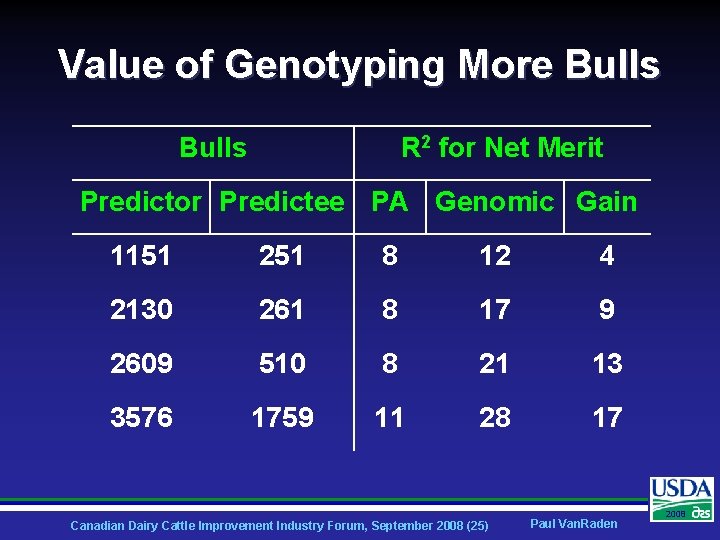
Value of Genotyping More Bulls R 2 for Net Merit Predictor Predictee PA Genomic Gain 1151 251 8 12 4 2130 261 8 17 9 2609 510 8 21 13 3576 1759 11 28 17 Canadian Dairy Cattle Improvement Industry Forum, September 2008 (25) Paul Van. Raden 2008

Simulated Results World Holstein Population Ø 15, 197 older and 5, 987 younger bulls in Interbull file Ø 40, 000 SNPs and 10, 000 QTLs Ø Provided timing, memory test Ø Reliability vs parent average REL • • • REL = corr 2 (EBV, true BV) 80% vs 34% expected for young bulls 72% vs 30% observed in simulation Canadian Dairy Cattle Improvement Industry Forum, September 2008 (26) Paul Van. Raden 2008

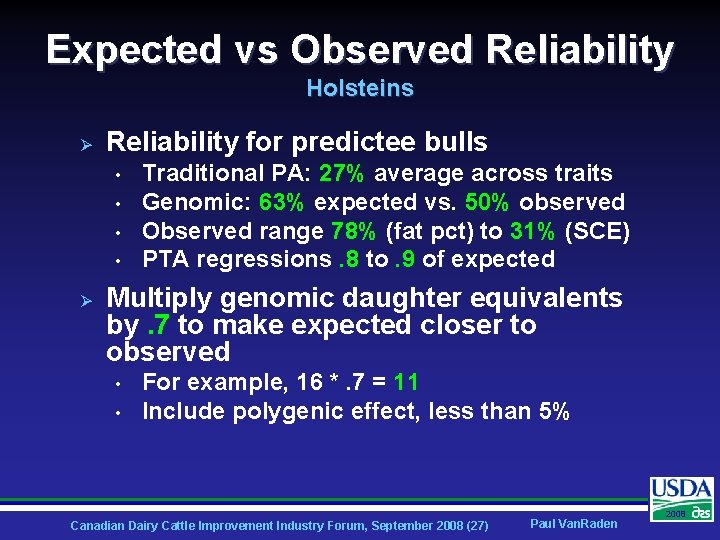
Expected vs Observed Reliability Holsteins Ø Reliability for predictee bulls • • Ø Traditional PA: 27% average across traits Genomic: 63% expected vs. 50% observed Observed range 78% (fat pct) to 31% (SCE) PTA regressions. 8 to. 9 of expected Multiply genomic daughter equivalents by. 7 to make expected closer to observed • • For example, 16 *. 7 = 11 Include polygenic effect, less than 5% Canadian Dairy Cattle Improvement Industry Forum, September 2008 (27) Paul Van. Raden 2008

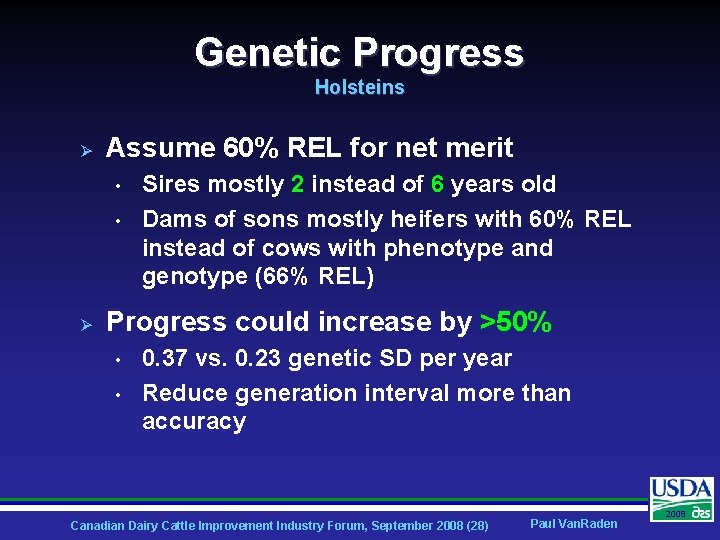
Genetic Progress Holsteins Ø Assume 60% REL for net merit • • Ø Sires mostly 2 instead of 6 years old Dams of sons mostly heifers with 60% REL instead of cows with phenotype and genotype (66% REL) Progress could increase by >50% • • 0. 37 vs. 0. 23 genetic SD per year Reduce generation interval more than accuracy Canadian Dairy Cattle Improvement Industry Forum, September 2008 (28) Paul Van. Raden 2008

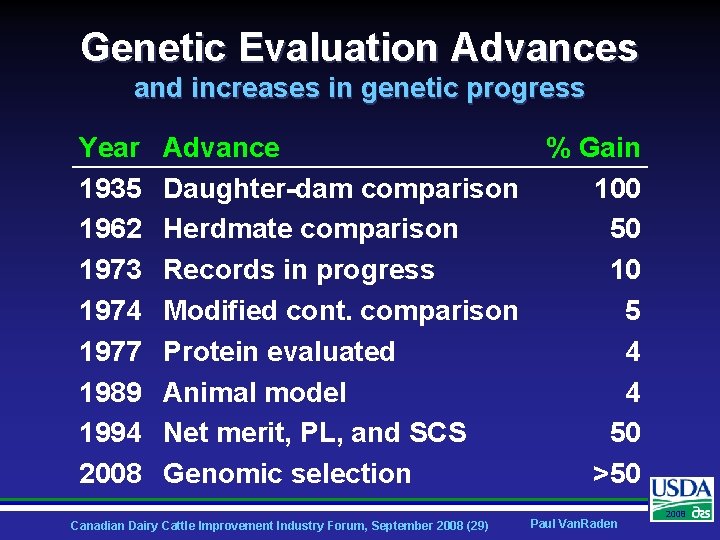
Genetic Evaluation Advances and increases in genetic progress Year 1935 1962 1973 1974 1977 1989 1994 2008 Advance % Gain Daughter-dam comparison 100 Herdmate comparison 50 Records in progress 10 Modified cont. comparison 5 Protein evaluated 4 Animal model 4 Net merit, PL, and SCS 50 Genomic selection >50 Canadian Dairy Cattle Improvement Industry Forum, September 2008 (29) Paul Van. Raden 2008

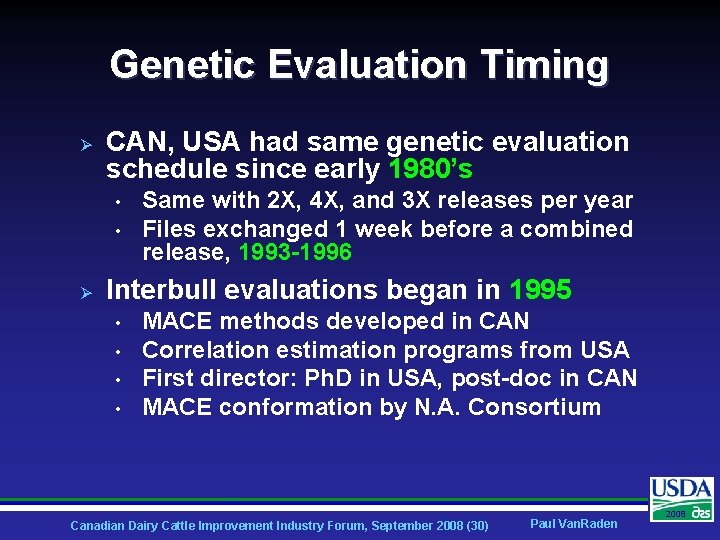
Genetic Evaluation Timing Ø CAN, USA had same genetic evaluation schedule since early 1980’s • • Ø Same with 2 X, 4 X, and 3 X releases per year Files exchanged 1 week before a combined release, 1993 -1996 Interbull evaluations began in 1995 • • MACE methods developed in CAN Correlation estimation programs from USA First director: Ph. D in USA, post-doc in CAN MACE conformation by N. A. Consortium Canadian Dairy Cattle Improvement Industry Forum, September 2008 (30) Paul Van. Raden 2008

CAN, USA Combined Data Ø Evaluations tested and reported in 1991 • • Ø Ø Banos and Wiggans, Robinson and Wiggans, Powell et al, Wiggans et al Both countries used Cornell computer Animal models applied to yield data of Jerseys and Ayrshires Correlations. 98 and. 96 between combined vs. converted evaluation Canadian Dairy Cattle Improvement Industry Forum, September 2008 (31) Paul Van. Raden 2008

Conclusions Ø Ø Ø Genomic predictions significantly better than parent average (P <. 0001) for all 26 traits tested High reliability requires many genotypes and phenotypes Close cooperation between CAN and USA will continue • Science, software, and data sharing Canadian Dairy Cattle Improvement Industry Forum, September 2008 (32) Paul Van. Raden 2008

Acknowledgments Ø Genotyping and DNA extraction: • Ø Computing: • Ø BFGL, U. Missouri, U. Alberta, Gene. Seek, GIFV, and Illumina AIPL staff (Mel Tooker, Leigh Walton, etc. ) Funding: • National Research Initiative grants – • • 2006 -35205 -16888, 2006 -35205 -16701 Agriculture Research Service Contributors to Cooperative Dairy DNA Repository (CDDR) Canadian Dairy Cattle Improvement Industry Forum, September 2008 (33) Paul Van. Raden 2008

CDDR Contributors Ø National Association of Animal Breeders (NAAB, Columbia, MO) • ABS Global (De. Forest, WI) Accelerated Genetics (Baraboo, WI) Alta (Balzac, AB) Genex (Shawano, WI) New Generation Genetics (Fort Atkinson, WI) • Select Sires (Plain City, OH) • Semex Alliance (Guelph, ON) • Taurus-Service (Mehoopany, PA) • • Canadian Dairy Cattle Improvement Industry Forum, September 2008 (34) Paul Van. Raden 2008
 Inter american accreditation cooperation
Inter american accreditation cooperation Principle of genomic equivalence
Principle of genomic equivalence Genomic england
Genomic england Genomic england
Genomic england Genomic england
Genomic england Genomic instability
Genomic instability Genomic
Genomic Genomic imprinting definition
Genomic imprinting definition Genomic signal processing
Genomic signal processing Comparative genomic hybridization animation
Comparative genomic hybridization animation Genomic equivalence definition
Genomic equivalence definition True north vs magnetic north
True north vs magnetic north North east and north cumbria ics map
North east and north cumbria ics map Chapter 14 lesson 1 the industrial north
Chapter 14 lesson 1 the industrial north The north pole ____ a latitude of 90 degrees north
The north pole ____ a latitude of 90 degrees north North american air masses
North american air masses Color the north america biomes
Color the north america biomes North american regional reanalysis
North american regional reanalysis Air masses and fronts
Air masses and fronts Who maintains the north american cargo securement standard
Who maintains the north american cargo securement standard North american gaming regulators association
North american gaming regulators association North american association for environmental education
North american association for environmental education Association of north american missions
Association of north american missions Namb
Namb Laissez faire theory
Laissez faire theory North american plate
North american plate Precambrian animals
Precambrian animals Fmcsa north american standard level i course online
Fmcsa north american standard level i course online North american portal
North american portal North american free trade agreement
North american free trade agreement North american colonies
North american colonies North american carbon program
North american carbon program What does cooperation look like
What does cooperation look like Example of cooperation in social interaction
Example of cooperation in social interaction International laboratory accreditation cooperation (ilac)
International laboratory accreditation cooperation (ilac)