Epigenetics Genomic imprinting Genomic Imprinting Preferential expression or




- Slides: 13

Epigenetics: Genomic imprinting

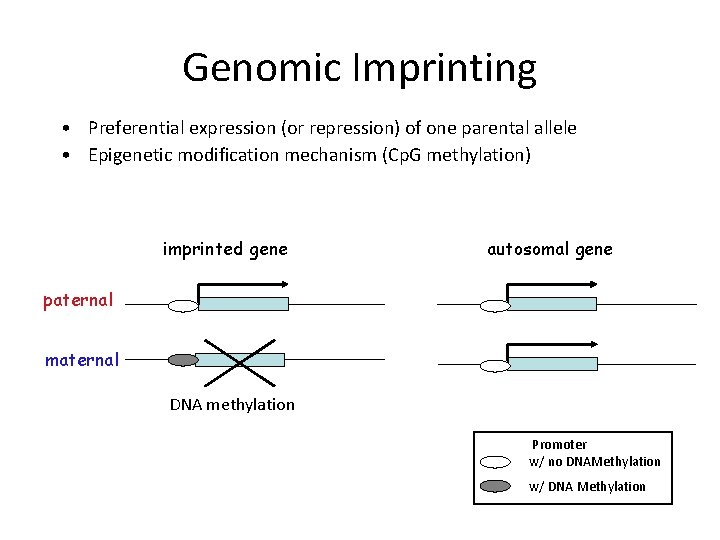
Genomic Imprinting • Preferential expression (or repression) of one parental allele • Epigenetic modification mechanism (Cp. G methylation) imprinted gene autosomal gene paternal maternal DNA methylation Promoter w/ no DNAMethylation w/ DNA Methylation

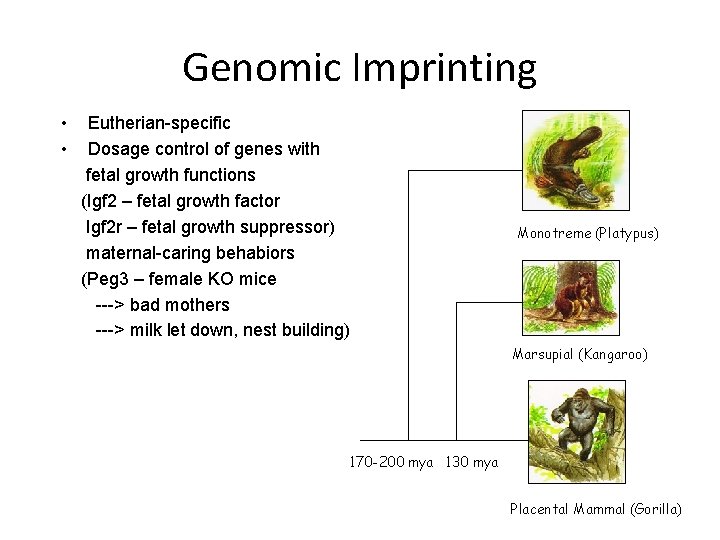
Genomic Imprinting • • Eutherian-specific Dosage control of genes with fetal growth functions (Igf 2 – fetal growth factor Igf 2 r – fetal growth suppressor) maternal-caring behabiors (Peg 3 – female KO mice ---> bad mothers ---> milk let down, nest building) Monotreme (Platypus) Marsupial (Kangaroo) 170 -200 mya 130 mya Placental Mammal (Gorilla)

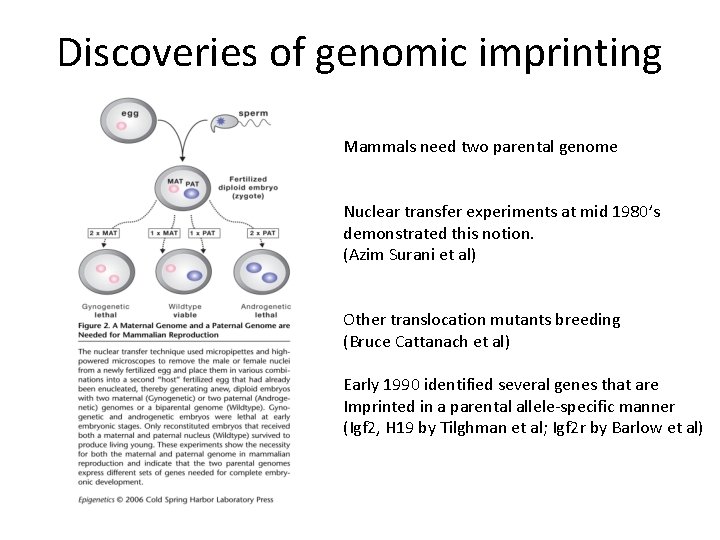
Discoveries of genomic imprinting Mammals need two parental genome Nuclear transfer experiments at mid 1980’s demonstrated this notion. (Azim Surani et al) Other translocation mutants breeding (Bruce Cattanach et al) Early 1990 identified several genes that are Imprinted in a parental allele-specific manner (Igf 2, H 19 by Tilghman et al; Igf 2 r by Barlow et al)

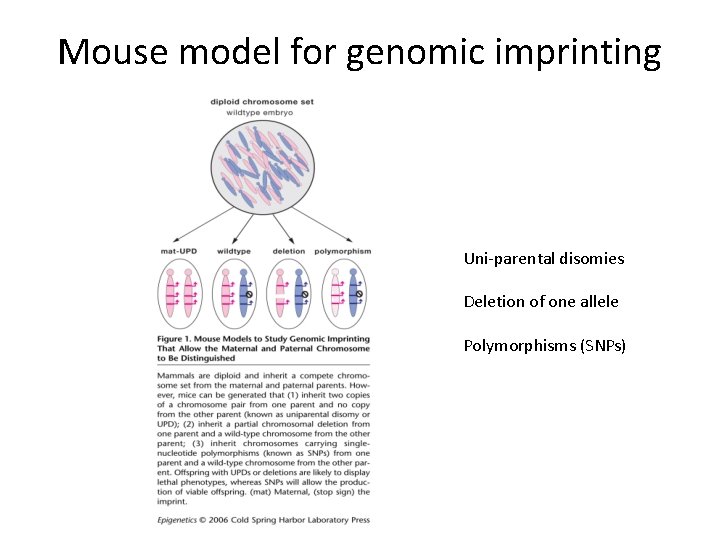
Mouse model for genomic imprinting Uni-parental disomies Deletion of one allele Polymorphisms (SNPs)

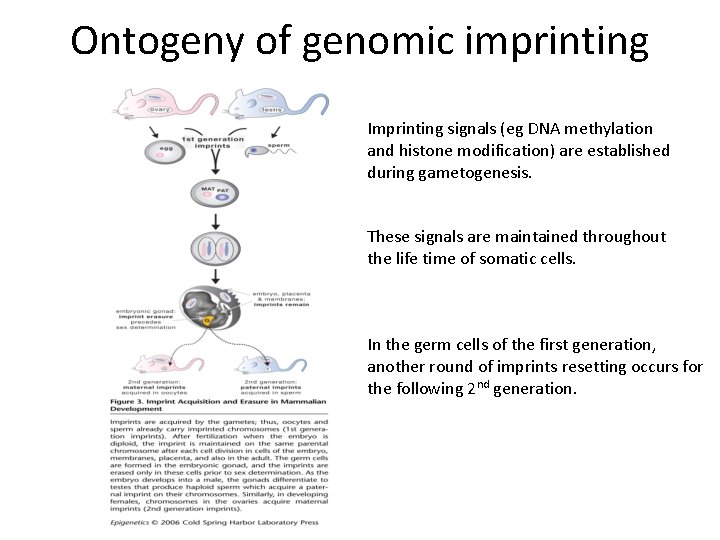
Ontogeny of genomic imprinting Imprinting signals (eg DNA methylation and histone modification) are established during gametogenesis. These signals are maintained throughout the life time of somatic cells. In the germ cells of the first generation, another round of imprints resetting occurs for the following 2 nd generation.

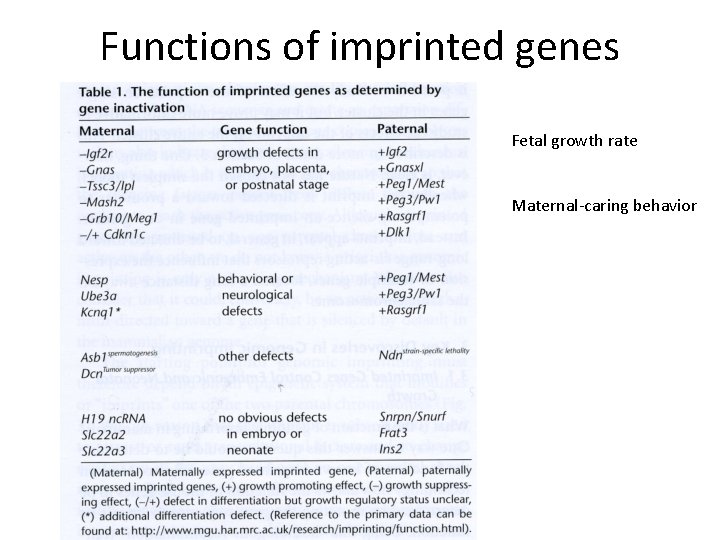
Functions of imprinted genes Fetal growth rate Maternal-caring behavior

Features of imprinted gene clusters • Imprinted genes are clustered in chromosomal regions. • Imprinted genes with both parental specificity are co-localized in a given domain. • Imprinted gene clusters tend to have long noncoding m. RNA genes, and these m. RNAs play significant roles for imprinting control. - Provide several paradigms for long distance transcriptional regulation in mammalian systems.

Features of imprinted gene clusters

Clustering of imprinted genes: one lnc (long non-coding) m. RNA + several protein-coding m. RNA

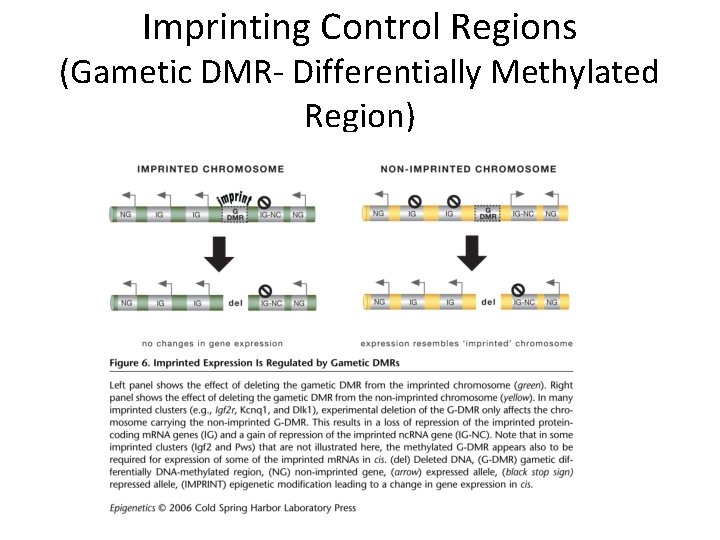
Imprinting Control Regions (Gametic DMR- Differentially Methylated Region)

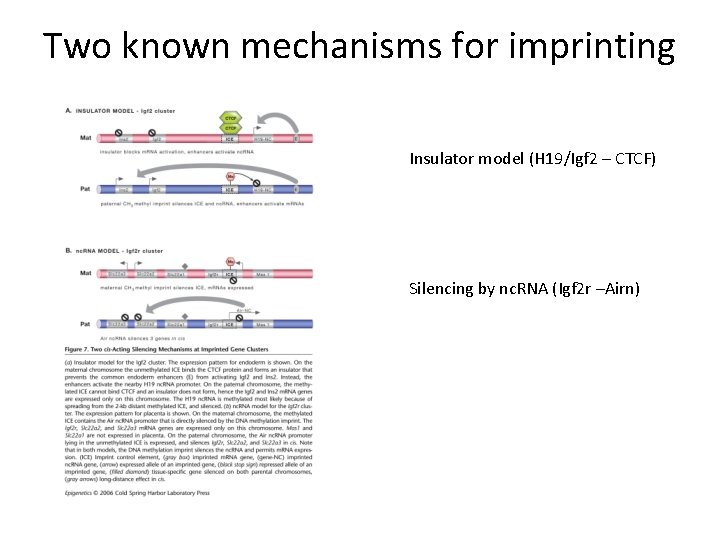
Two known mechanisms for imprinting Insulator model (H 19/Igf 2 – CTCF) Silencing by nc. RNA (Igf 2 r –Airn)

Unsolved questions (? ? ? ) • How ICRs (gametic DMRs) obtain DNA methylation during gametogenesis • How these DMRs are maintained throughout development, in particular early embryogenesis. • Each imprinted cluster has different mechanisms for allele-specific expression in somatic cells, which requires more thorough analyses.