Epigenetics Histone Modification I Nucleosome A packaging unit



- Slides: 15

Epigenetics: Histone Modification I

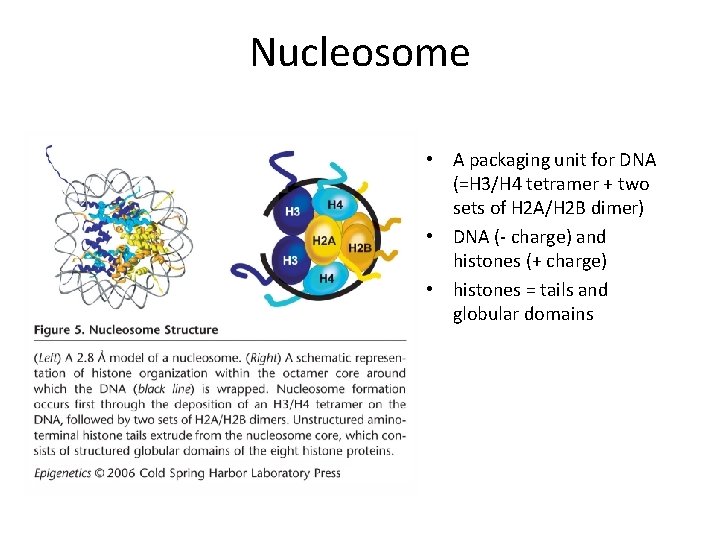
Nucleosome • A packaging unit for DNA (=H 3/H 4 tetramer + two sets of H 2 A/H 2 B dimer) • DNA (- charge) and histones (+ charge) • histones = tails and globular domains

Higher-Order Packaging of Chromatins • An efficient packaging strategy (10, 000 -fold compaction) • A barrier for gene transcription

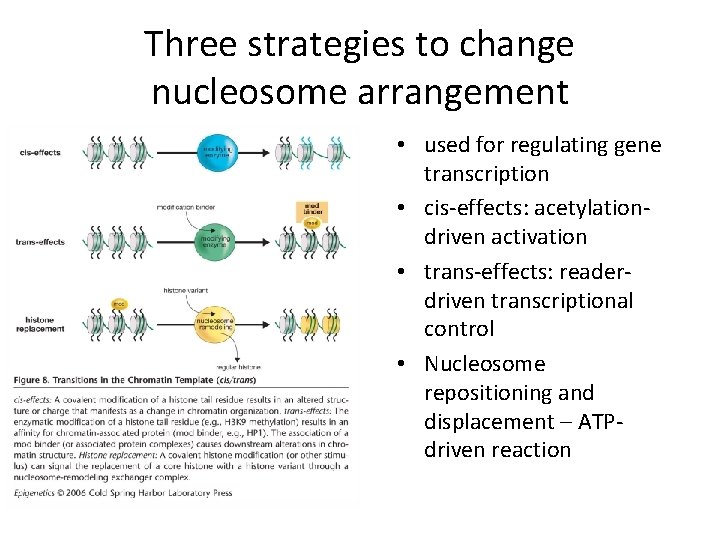
Three strategies to change nucleosome arrangement • used for regulating gene transcription • cis-effects: acetylationdriven activation • trans-effects: readerdriven transcriptional control • Nucleosome repositioning and displacement – ATPdriven reaction

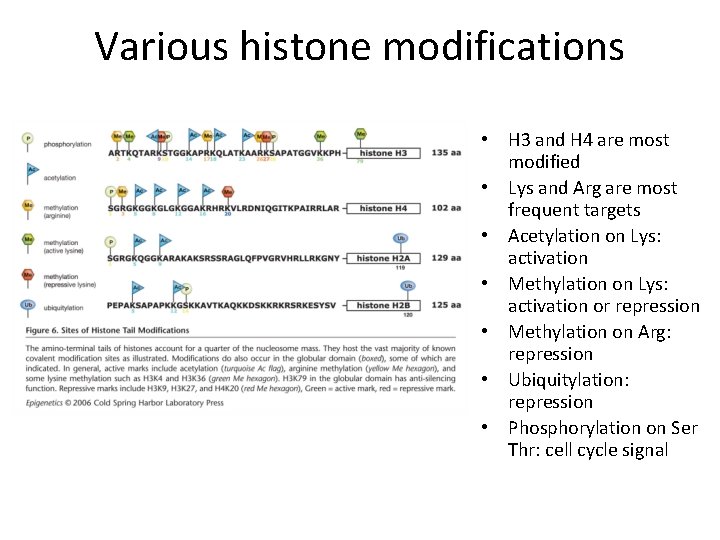
Various histone modifications • H 3 and H 4 are most modified • Lys and Arg are most frequent targets • Acetylation on Lys: activation • Methylation on Lys: activation or repression • Methylation on Arg: repression • Ubiquitylation: repression • Phosphorylation on Ser Thr: cell cycle signal

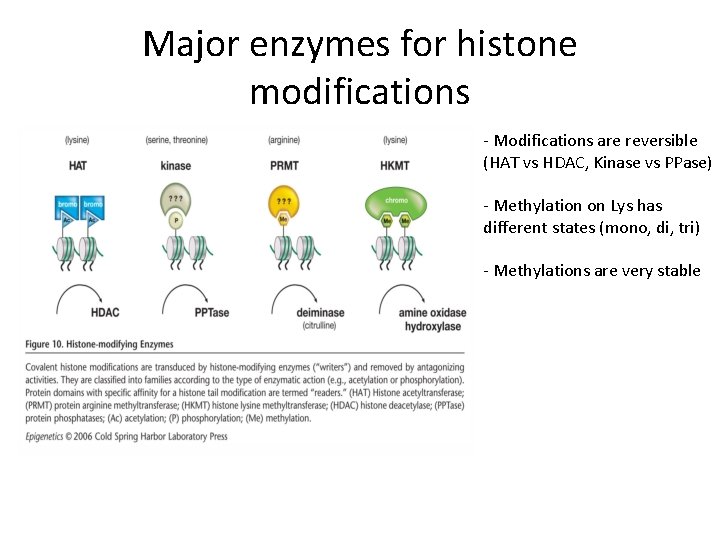
Major enzymes for histone modifications - Modifications are reversible (HAT vs HDAC, Kinase vs PPase) - Methylation on Lys has different states (mono, di, tri) - Methylations are very stable

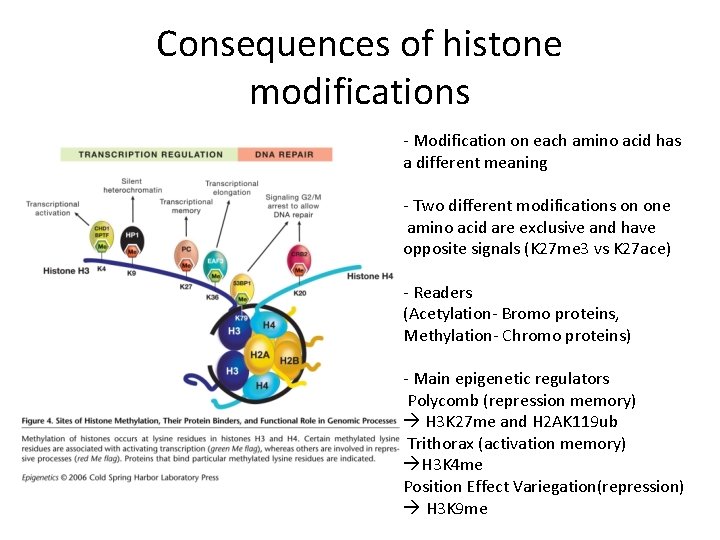
Consequences of histone modifications - Modification on each amino acid has a different meaning - Two different modifications on one amino acid are exclusive and have opposite signals (K 27 me 3 vs K 27 ace) - Readers (Acetylation- Bromo proteins, Methylation- Chromo proteins) - Main epigenetic regulators Polycomb (repression memory) H 3 K 27 me and H 2 AK 119 ub Trithorax (activation memory) H 3 K 4 me Position Effect Variegation(repression) H 3 K 9 me

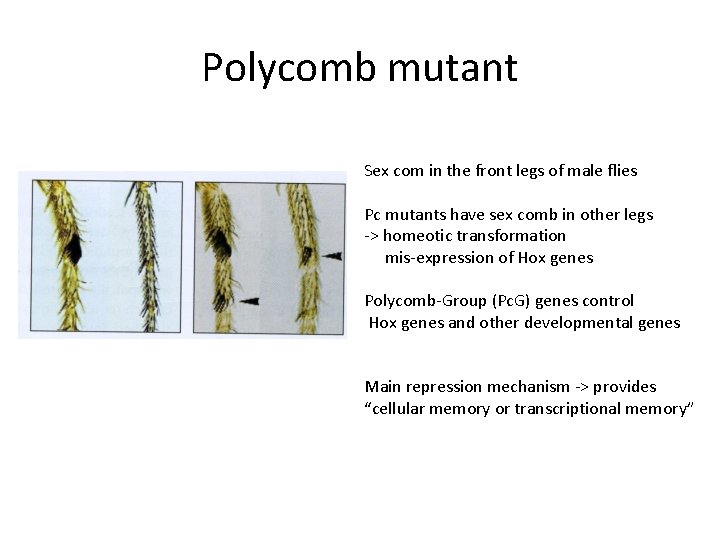
Polycomb mutant Sex com in the front legs of male flies Pc mutants have sex comb in other legs -> homeotic transformation mis-expression of Hox genes Polycomb-Group (Pc. G) genes control Hox genes and other developmental genes Main repression mechanism -> provides “cellular memory or transcriptional memory”

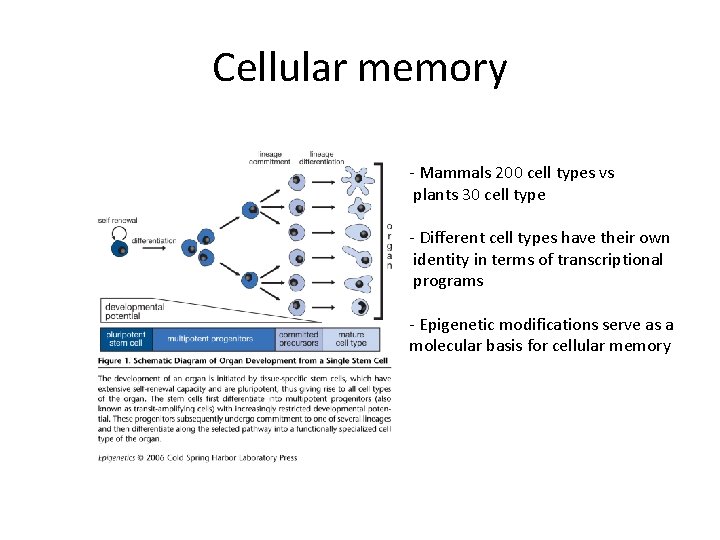
Cellular memory - Mammals 200 cell types vs plants 30 cell type - Different cell types have their own identity in terms of transcriptional programs - Epigenetic modifications serve as a molecular basis for cellular memory

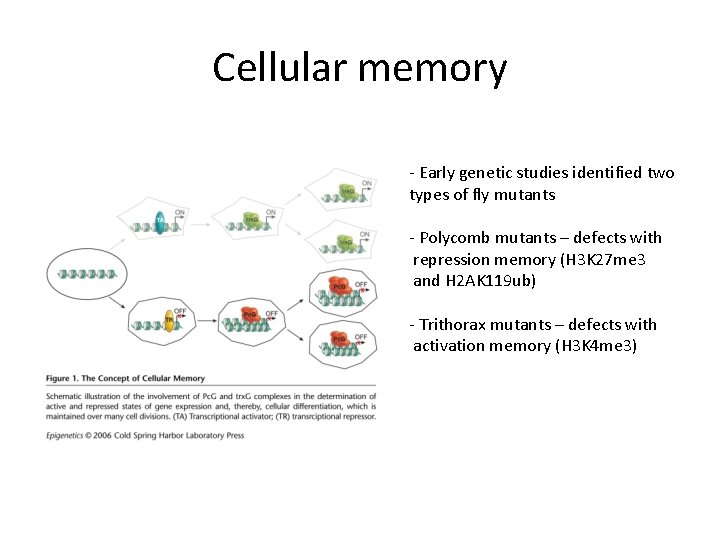
Cellular memory - Early genetic studies identified two types of fly mutants - Polycomb mutants – defects with repression memory (H 3 K 27 me 3 and H 2 AK 119 ub) - Trithorax mutants – defects with activation memory (H 3 K 4 me 3)

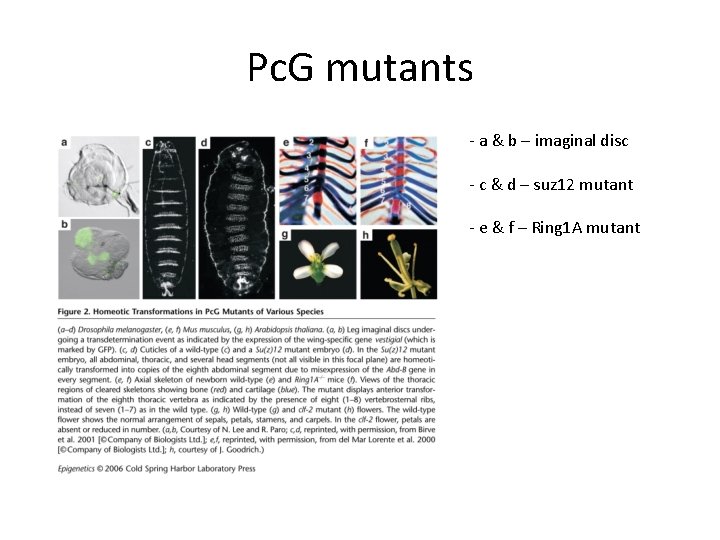
Pc. G mutants - a & b – imaginal disc - c & d – suz 12 mutant - e & f – Ring 1 A mutant

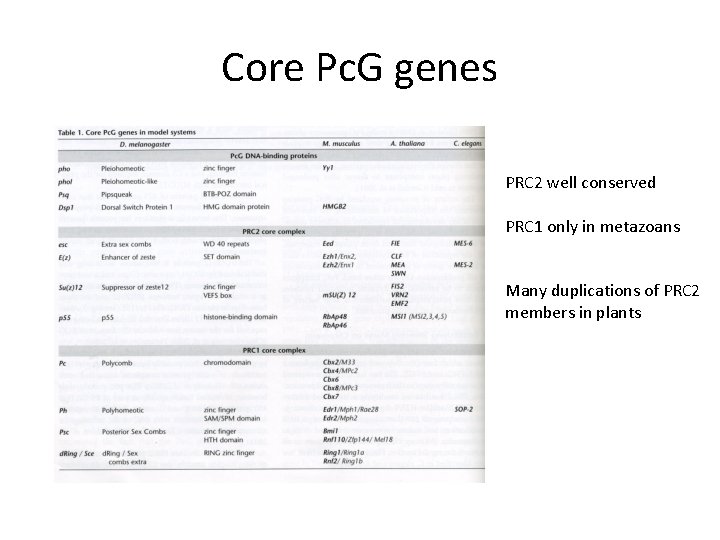
Core Pc. G genes PRC 2 well conserved PRC 1 only in metazoans Many duplications of PRC 2 members in plants

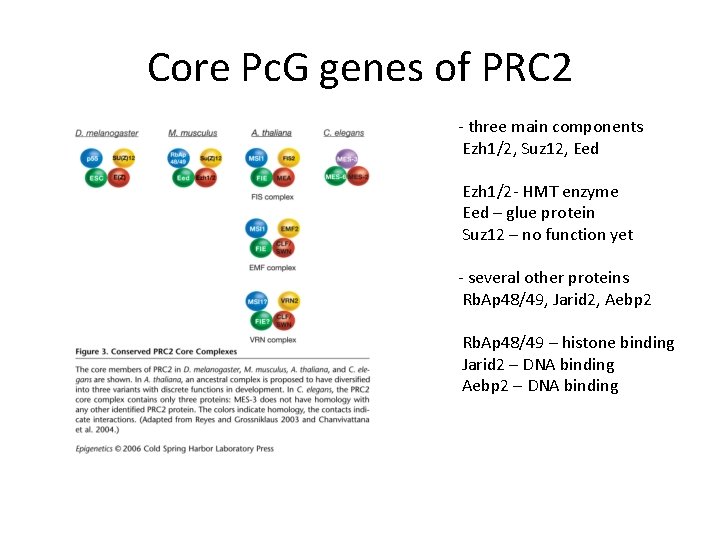
Core Pc. G genes of PRC 2 - three main components Ezh 1/2, Suz 12, Eed Ezh 1/2 - HMT enzyme Eed – glue protein Suz 12 – no function yet - several other proteins Rb. Ap 48/49, Jarid 2, Aebp 2 Rb. Ap 48/49 – histone binding Jarid 2 – DNA binding Aebp 2 – DNA binding

Pc. G-dependent repression PRC 2 -> H 3 K 27 -> PRC 1 -> H 2 AK 119 ub Targeting of PRC 2 & PRC 1 is mediated either by nc. RNA or DNA-bindig protein Maintenance of H 3 K 27 me 3 during DNA replication is mediated through Eed

Genome-wide PRE sites - more than Hox cluster - many genes with tissue-specific genes are controlled through PRC-mediated mechanisms - PRC 2 serves as temporary repression mechanism (poised state) in early stage embryo - modify Cp. G islands