Modeling Functional Genomics Datasets CVM 8890 101 Lesson






- Slides: 35

Modeling Functional Genomics Datasets CVM 8890 -101 Lesson 6 11 July 2007 Bindu Nanduri

Lesson 6: Functional genomics modeling II: a pathway analysis example.

Introduction to protein interaction networks


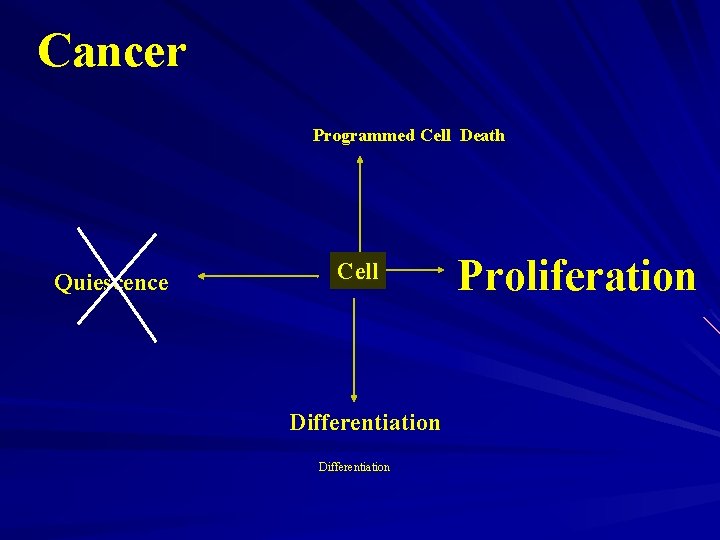
Cancer Programmed Cell Death Quiescence Cell Differentiation Proliferation

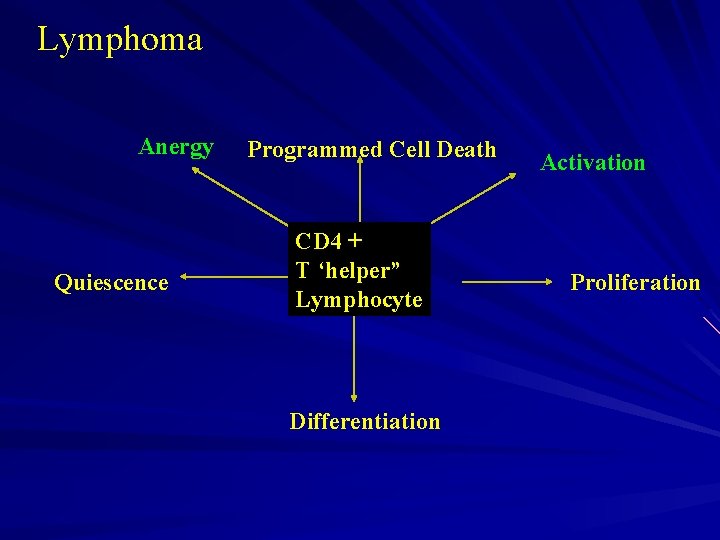
Lymphoma Anergy Quiescence Programmed Cell Death CD 4 + T ‘helper” Lymphocyte Differentiation Activation Proliferation

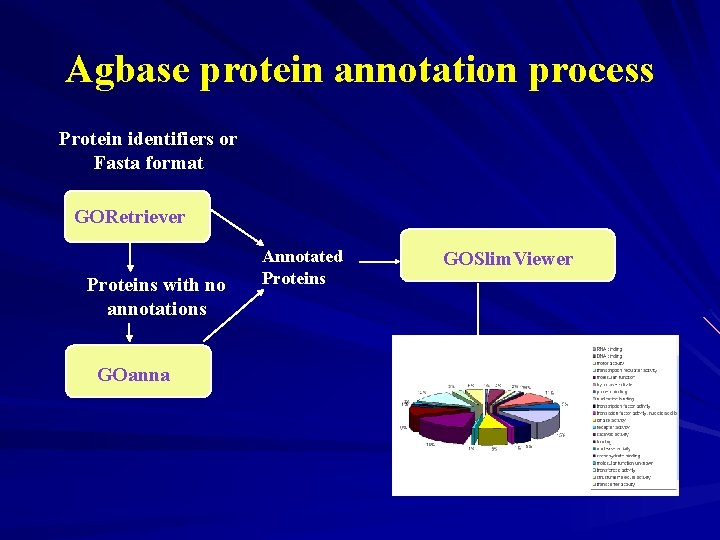
Agbase protein annotation process Protein identifiers or Fasta format GORetriever Proteins with no annotations GOanna Annotated Proteins GOSlim. Viewer


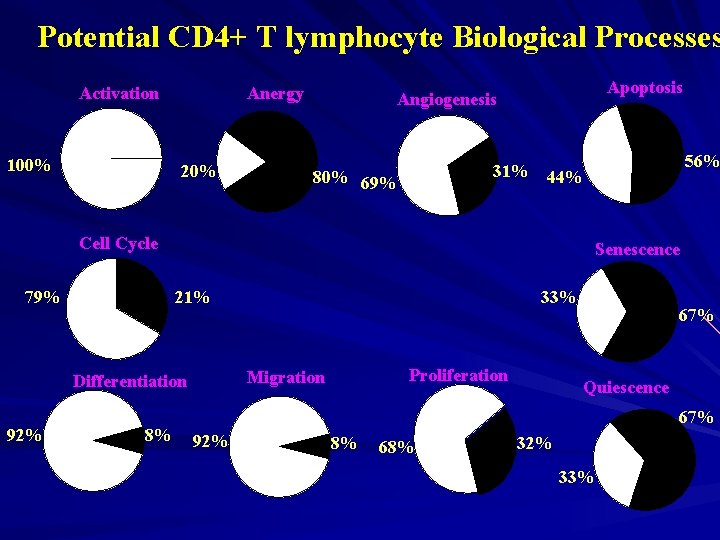
Potential CD 4+ T lymphocyte Biological Processes Activation 100% Anergy 20% Apoptosis Angiogenesis 31% 80% 69% 56% 44% Cell Cycle 79% Senescence 21% 8% Proliferation Migration Differentiation 92% 33% 67% Quiescence 67% 92% 8% 68% 32% 33%


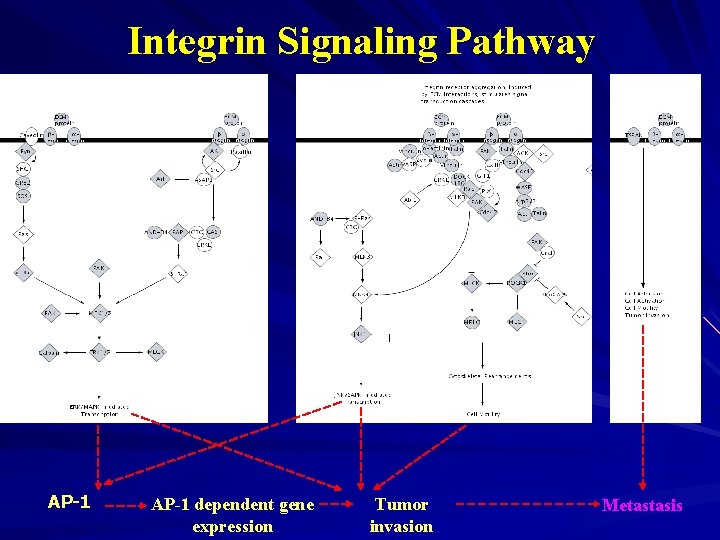
Integrin Signaling Pathway AP-1 dependent gene expression Tumor invasion Metastasis

Hypothesis driven data analysis Exploration of data to identify pathways of interacting proteins Protein protein interaction networks (PPI)

Why study PPIs Proteins do not function alone!!!!! PPI are inherent to the function of multiprotein complexes PPIs can help infer function : where functional information is available for one partner Changes in normal PPI can result in disease

Types of PPI

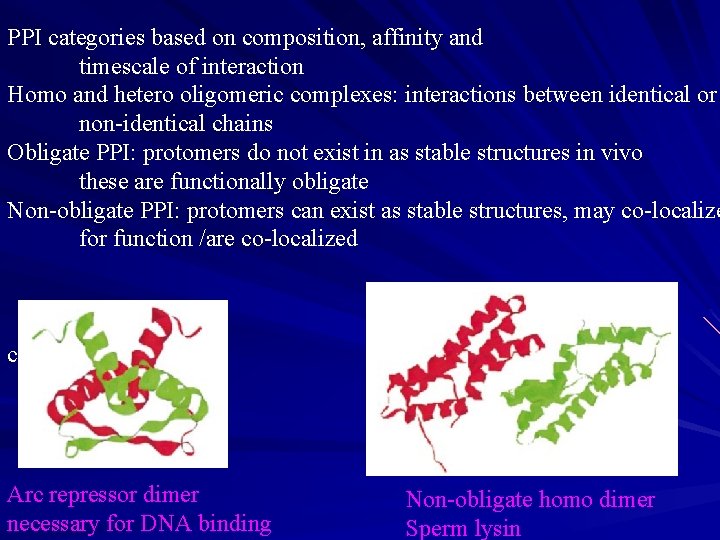
PPI categories based on composition, affinity and timescale of interaction Homo and hetero oligomeric complexes: interactions between identical or non-identical chains Obligate PPI: protomers do not exist in as stable structures in vivo these are functionally obligate Non-obligate PPI: protomers can exist as stable structures, may co-localize for function /are co-localized c Arc repressor dimer necessary for DNA binding Non-obligate homo dimer Sperm lysin

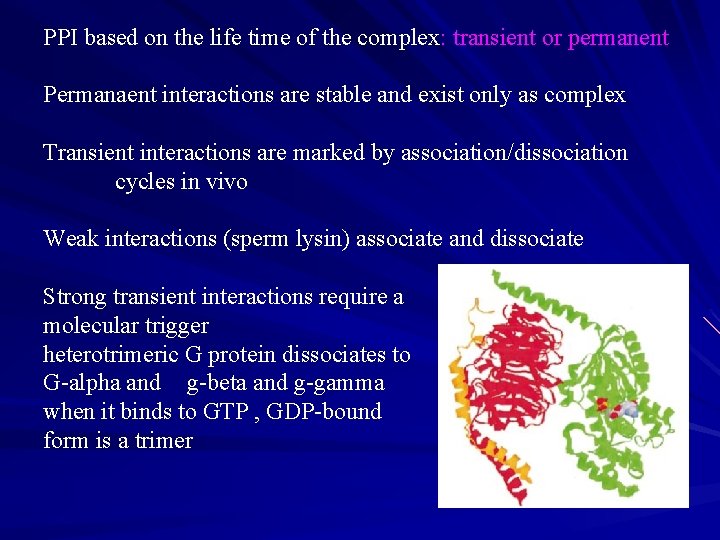
PPI based on the life time of the complex: transient or permanent Permanaent interactions are stable and exist only as complex Transient interactions are marked by association/dissociation cycles in vivo Weak interactions (sperm lysin) associate and dissociate Strong transient interactions require a molecular trigger heterotrimeric G protein dissociates to G-alpha and g-beta and g-gamma when it binds to GTP , GDP-bound form is a trimer

Control of protein oligomerization PPI interactions are a continuum of obligate and non-obligate states Interactions of complexes driven by concentration and free energy of complex relative to alternate states

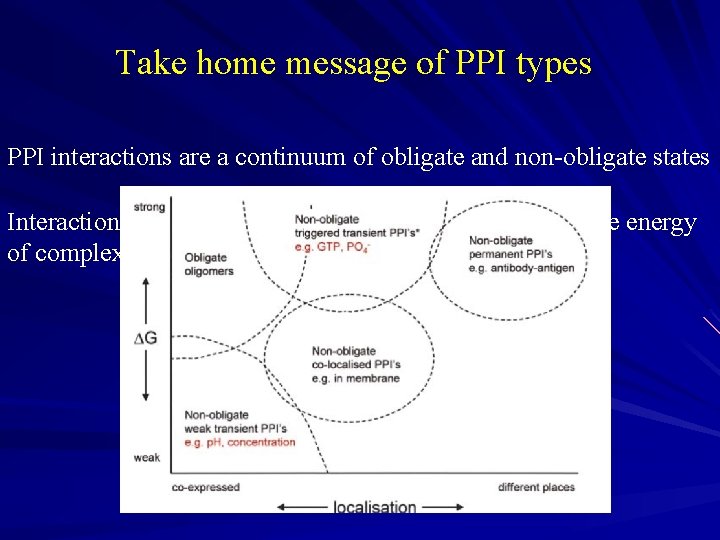
Take home message of PPI types PPI interactions are a continuum of obligate and non-obligate states Interactions of complexes driven by concentration and free energy of complex relative to alternate states

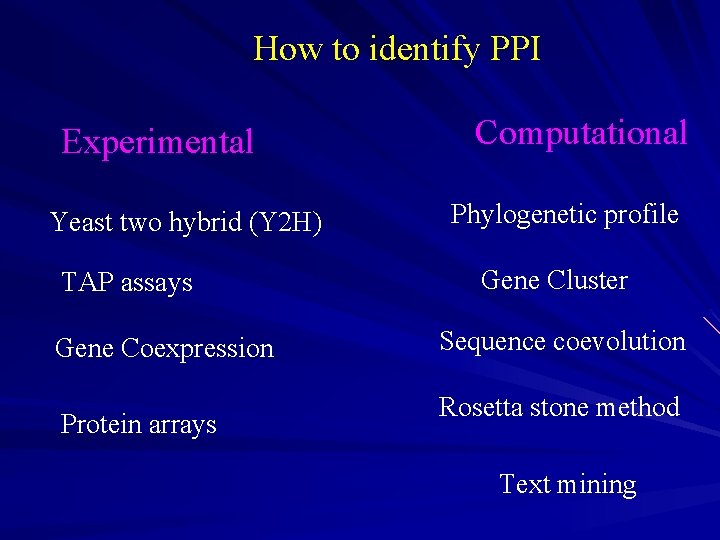
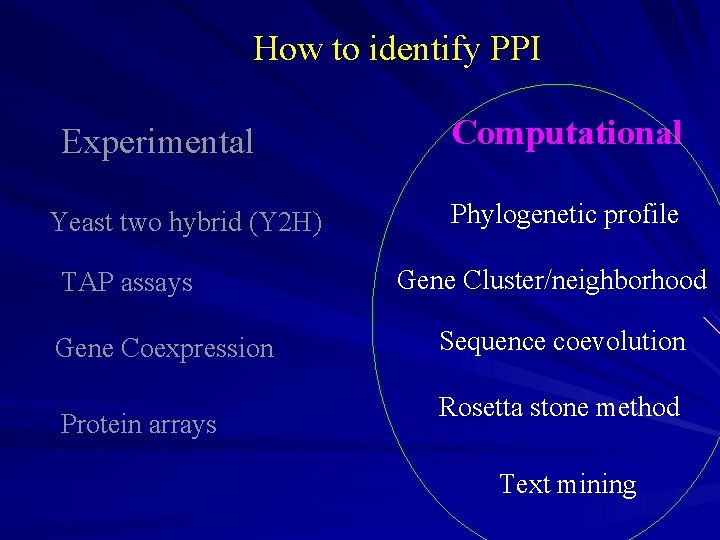
How to identify PPI Experimental Yeast two hybrid (Y 2 H) TAP assays Gene Coexpression Protein arrays Computational Phylogenetic profile Gene Cluster Sequence coevolution Rosetta stone method Text mining

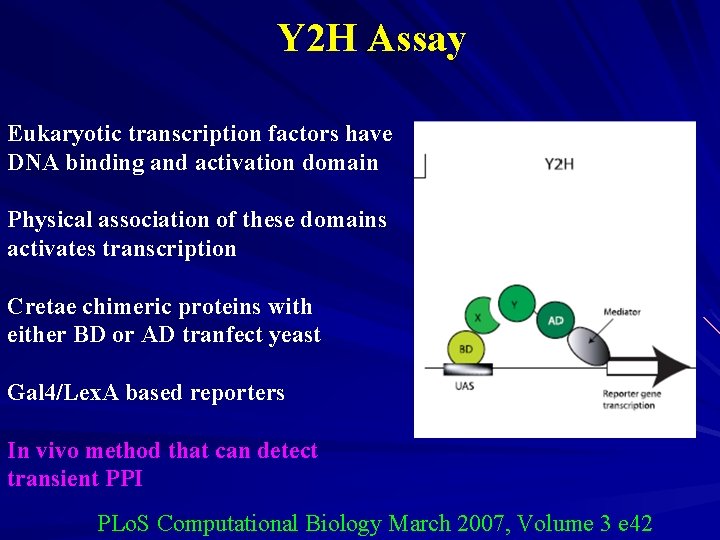
Y 2 H Assay Eukaryotic transcription factors have DNA binding and activation domain Physical association of these domains activates transcription Cretae chimeric proteins with either BD or AD tranfect yeast Gal 4/Lex. A based reporters In vivo method that can detect transient PPI PLo. S Computational Biology March 2007, Volume 3 e 42

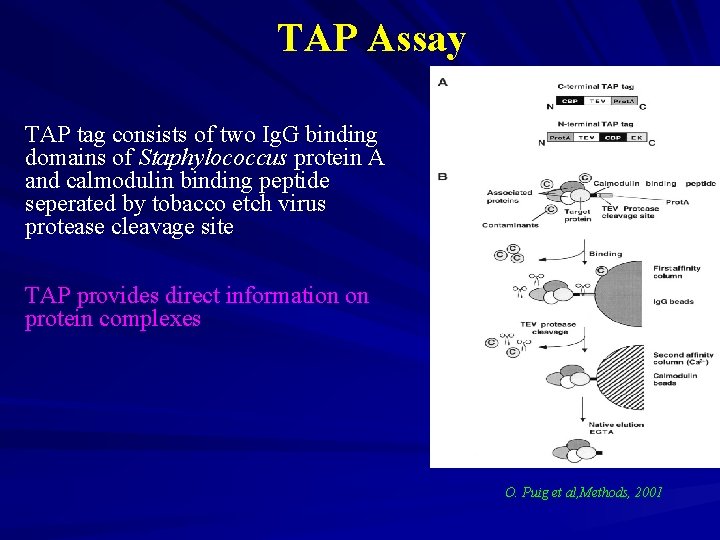
TAP Assay TAP tag consists of two Ig. G binding domains of Staphylococcus protein A and calmodulin binding peptide seperated by tobacco etch virus protease cleavage site TAP provides direct information on protein complexes O. Puig et al, Methods, 2001

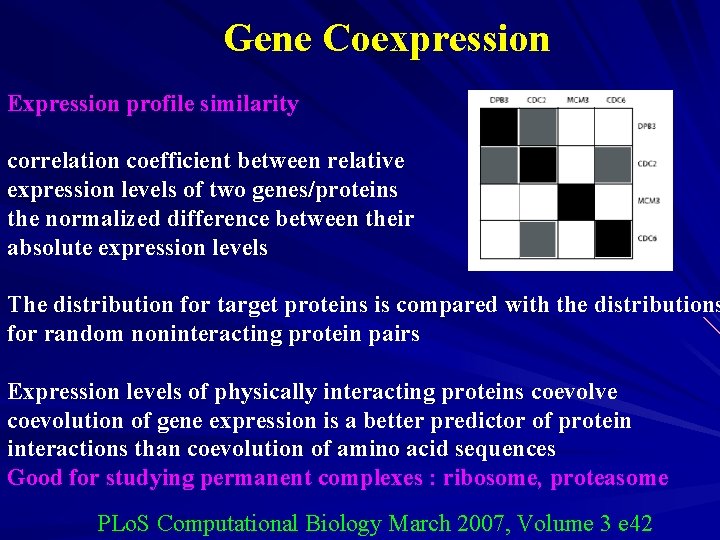
Gene Coexpression Expression profile similarity correlation coefficient between relative expression levels of two genes/proteins the normalized difference between their absolute expression levels The distribution for target proteins is compared with the distributions for random noninteracting protein pairs Expression levels of physically interacting proteins coevolve coevolution of gene expression is a better predictor of protein interactions than coevolution of amino acid sequences Good for studying permanent complexes : ribosome, proteasome PLo. S Computational Biology March 2007, Volume 3 e 42

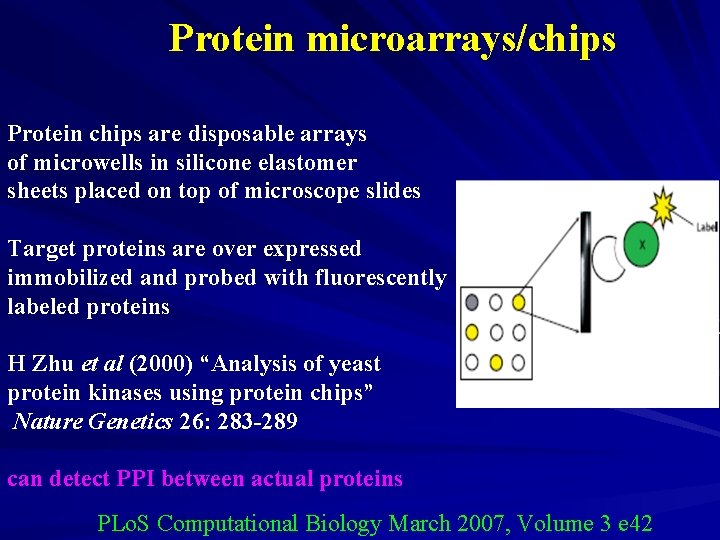
Protein microarrays/chips Protein chips are disposable arrays of microwells in silicone elastomer sheets placed on top of microscope slides Target proteins are over expressed immobilized and probed with fluorescently labeled proteins H Zhu et al (2000) “Analysis of yeast protein kinases using protein chips” Nature Genetics 26: 283 -289 can detect PPI between actual proteins PLo. S Computational Biology March 2007, Volume 3 e 42

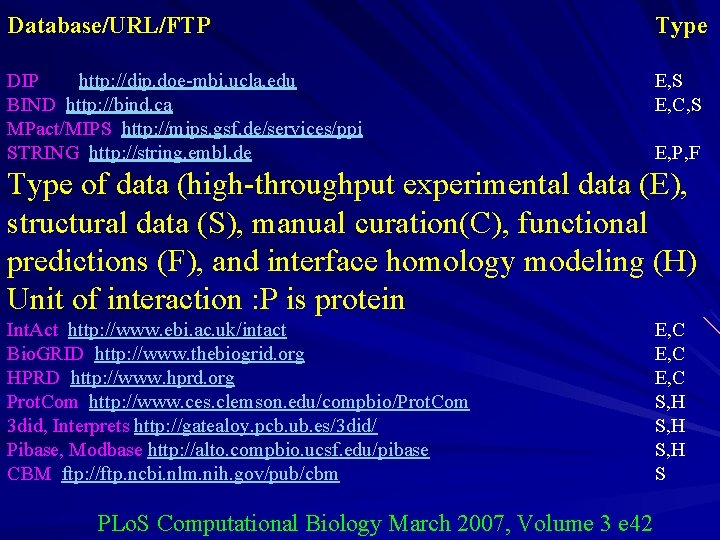
Database/URL/FTP Type DIP http: //dip. doe-mbi. ucla. edu BIND http: //bind. ca MPact/MIPS http: //mips. gsf. de/services/ppi STRING http: //string. embl. de MINT http: //mint. bio. uniroma 2. it/mint Int. Act http: //www. ebi. ac. uk/intact Bio. GRID http: //www. thebiogrid. org HPRD http: //www. hprd. org Prot. Com http: //www. ces. clemson. edu/compbio/Prot. Com 3 did, Interprets http: //gatealoy. pcb. ub. es/3 did/ Pibase, Modbase http: //alto. compbio. ucsf. edu/pibase CBM ftp: //ftp. ncbi. nlm. nih. gov/pub/cbm SCOPPI http: //www. scoppi. org/ i. Pfam http: //www. sanger. ac. uk/Software/Pfam/i. Pfam Inter. Dom http: //interdom. lit. org. sg DIMA http: //mips. gsf. de/genre/proj/dima/index. html Prolinks http: //prolinks. doe-mbi. ucla. edu/cgibin/functionator/pronav/ Predictome http: //predictome. bu. edu/ E, S E, C, S PLo. S Computational Biology March 2007, Volume 3 e 42 E, P, F E, C S, H S S P F, S F F

Database/URL/FTP Type DIP http: //dip. doe-mbi. ucla. edu BIND http: //bind. ca MPact/MIPS http: //mips. gsf. de/services/ppi STRING http: //string. embl. de E, S E, C, S E, P, F Type of data (high-throughput experimental data (E), structural data (S), manual curation(C), functional predictions (F), and interface homology modeling (H) Unit of interaction : P is protein Int. Act http: //www. ebi. ac. uk/intact Bio. GRID http: //www. thebiogrid. org HPRD http: //www. hprd. org Prot. Com http: //www. ces. clemson. edu/compbio/Prot. Com 3 did, Interprets http: //gatealoy. pcb. ub. es/3 did/ Pibase, Modbase http: //alto. compbio. ucsf. edu/pibase CBM ftp: //ftp. ncbi. nlm. nih. gov/pub/cbm PLo. S Computational Biology March 2007, Volume 3 e 42 E, C S, H S

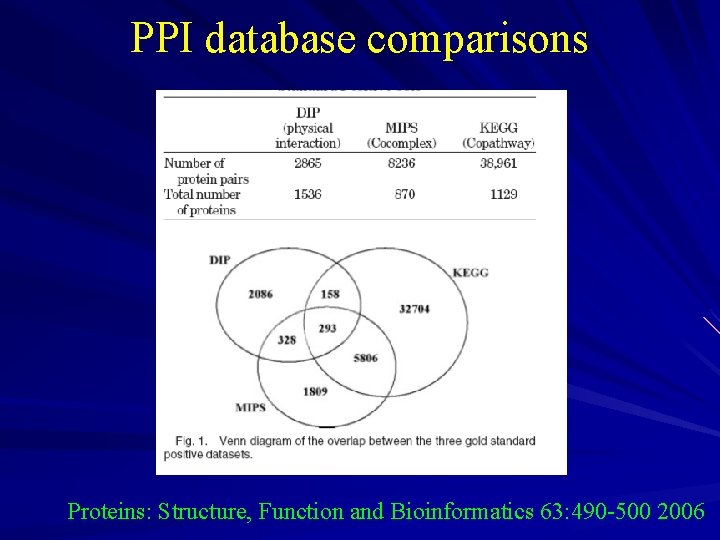
PPI database comparisons Proteins: Structure, Function and Bioinformatics 63: 490 -500 2006

Experimental PPI dataset overlap is small High FP rate in high- throughput exp ……. difficult to confirm by multiple sources

How to identify PPI Experimental Yeast two hybrid (Y 2 H) TAP assays Gene Coexpression Protein arrays Computational Phylogenetic profile Gene Cluster/neighborhood Sequence coevolution Rosetta stone method Text mining

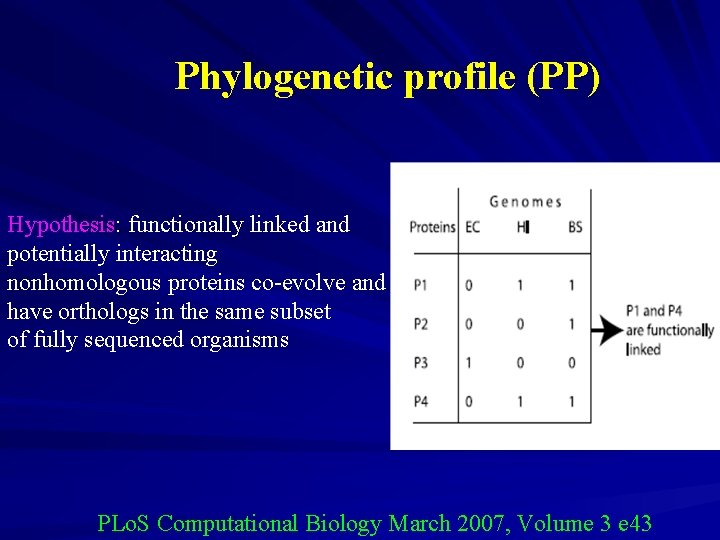
Phylogenetic profile (PP) Hypothesis: functionally linked and potentially interacting nonhomologous proteins co-evolve and have orthologs in the same subset of fully sequenced organisms PLo. S Computational Biology March 2007, Volume 3 e 43

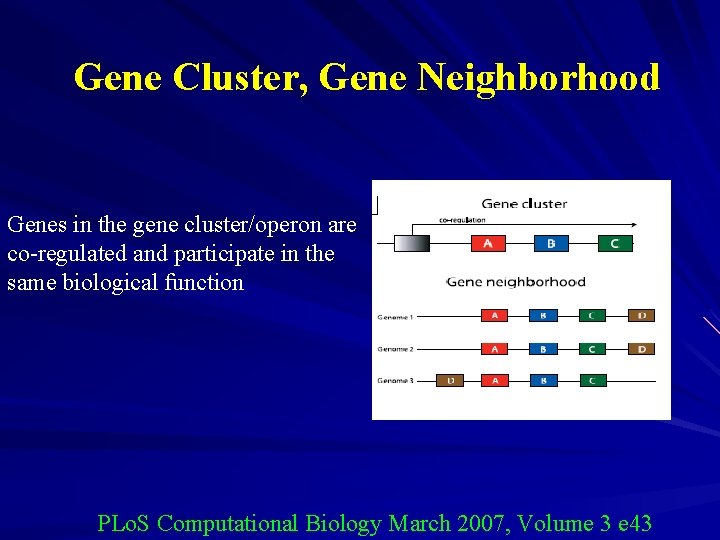
Gene Cluster, Gene Neighborhood Genes in the gene cluster/operon are co-regulated and participate in the same biological function PLo. S Computational Biology March 2007, Volume 3 e 43

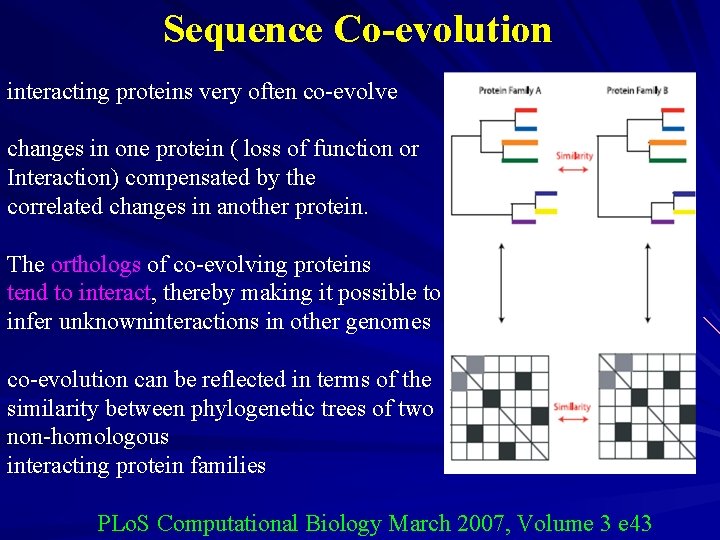
Sequence Co-evolution interacting proteins very often co-evolve changes in one protein ( loss of function or Interaction) compensated by the correlated changes in another protein. The orthologs of co-evolving proteins tend to interact, thereby making it possible to infer unknowninteractions in other genomes co-evolution can be reflected in terms of the similarity between phylogenetic trees of two non-homologous interacting protein families PLo. S Computational Biology March 2007, Volume 3 e 43

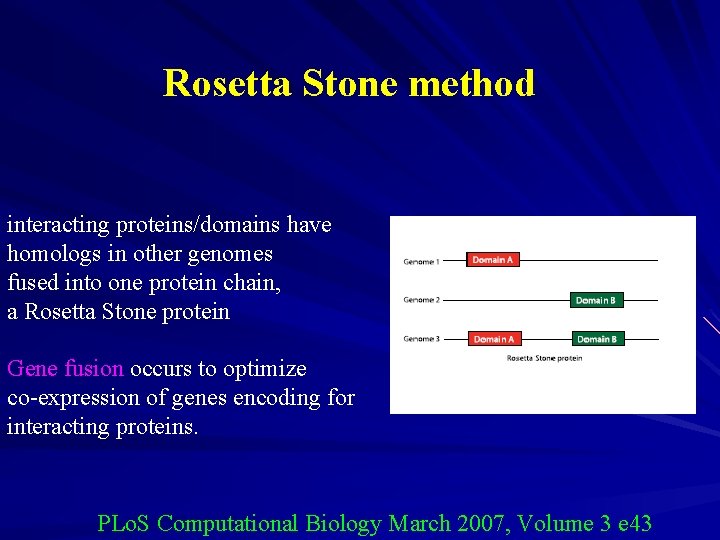
Rosetta Stone method interacting proteins/domains have homologs in other genomes fused into one protein chain, a Rosetta Stone protein Gene fusion occurs to optimize co-expression of genes encoding for interacting proteins. PLo. S Computational Biology March 2007, Volume 3 e 43

Text Mining Utilizing the wealth of publicly available data. . search Medline or Pub. Med for words or word combinations co-occurrence of words together is a simple metric, however prone to high false positive rates Natural Language Processing (NLP) methods are specific “A binds to B”; “A interacts with B”; “A associates with B” difficult to detect so it has a higher false negative rate Normally requires a list of known gene names or protein names for a given organism

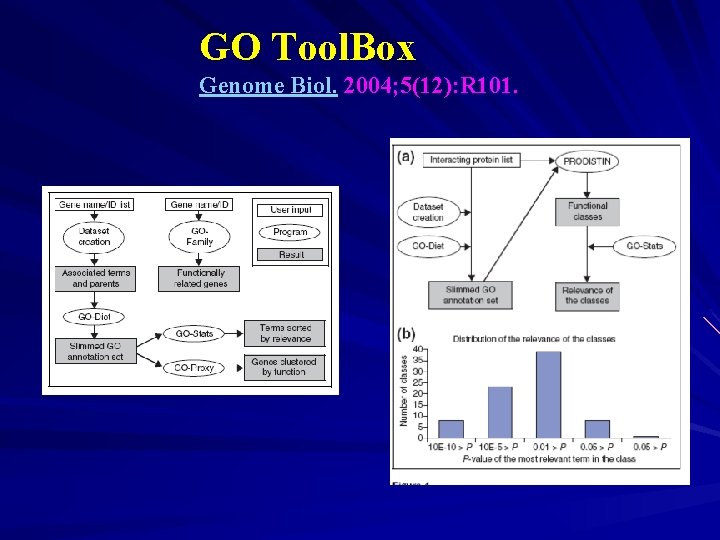
GO Tool. Box Genome Biol. 2004; 5(12): R 101.

Prot. Quant tool