Adding GO for Large Datasets COST Functional Modeling



- Slides: 13

Adding GO for Large Datasets COST Functional Modeling Workshop 22 -24 April, Helsinki

Large Datasets RNASeq data sets and etc. : § large data sets § often there is little functional information available § many enrichment analysis tools will not accept large gene lists § RNASeq data sets also contain “novel” genes

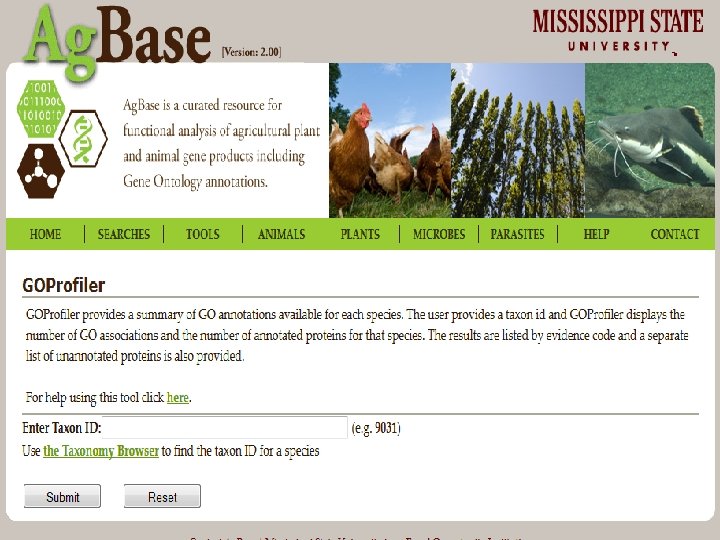
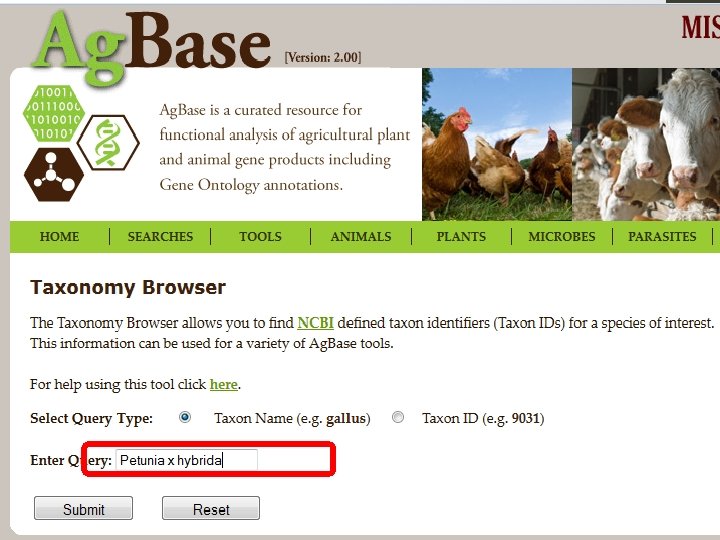
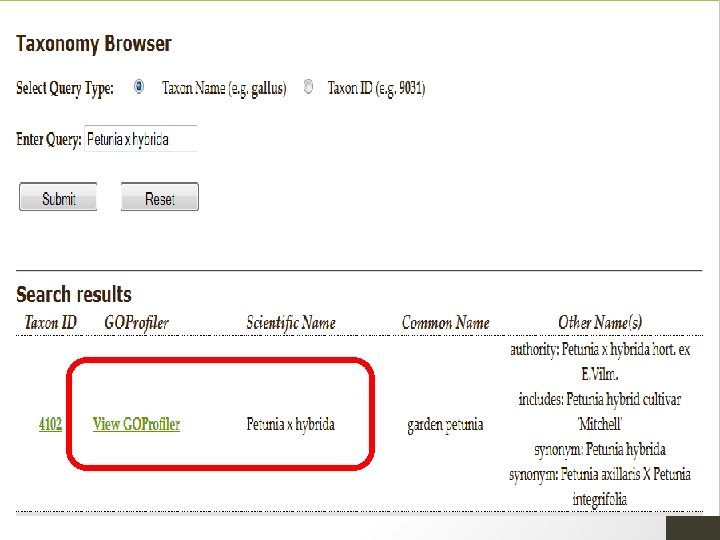
1. Finding Existing GO 1. Use GOProfiler to search based upon taxon or name. 2. Check the GO Consortium Website to see if your species of interest has an active annotation effort. • or to determine which relate species may have GO annotations that can be transferred 3. Use Quick. GO or GOProfiler to download existing GO annotations. 4. Add your own GO annotations…




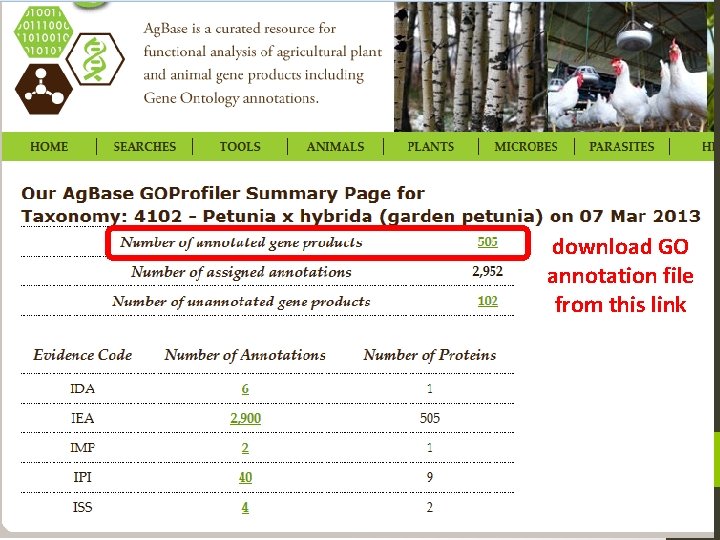
download GO annotation file from this link

http: //geneontology. org/

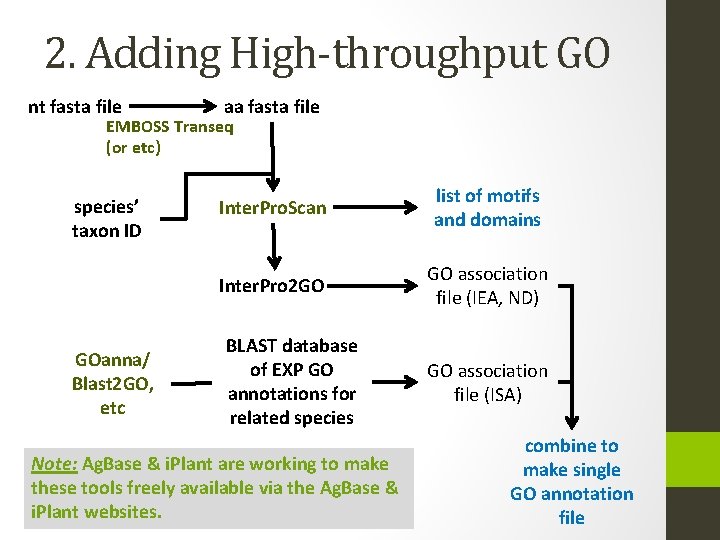
2. Adding High-throughput GO nt fasta file aa fasta file EMBOSS Transeq (or etc) species’ taxon ID GOanna/ Blast 2 GO, etc Inter. Pro. Scan list of motifs and domains Inter. Pro 2 GO GO association file (IEA, ND) BLAST database of EXP GO annotations for related species Note: Ag. Base & i. Plant are working to make these tools freely available via the Ag. Base & i. Plant websites. GO association file (ISA) combine to make single GO annotation file

http: //www. ebi. ac. uk/Tools/emboss

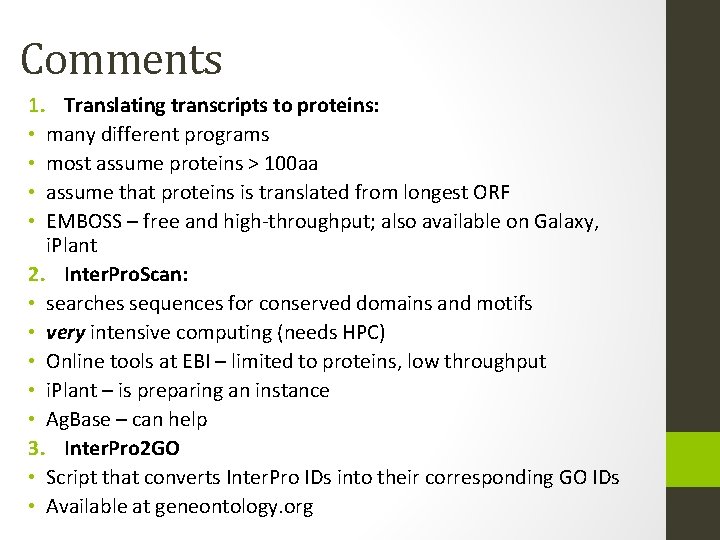
Comments 1. Translating transcripts to proteins: • many different programs • most assume proteins > 100 aa • assume that proteins is translated from longest ORF • EMBOSS – free and high-throughput; also available on Galaxy, i. Plant 2. Inter. Pro. Scan: • searches sequences for conserved domains and motifs • very intensive computing (needs HPC) • Online tools at EBI – limited to proteins, low throughput • i. Plant – is preparing an instance • Ag. Base – can help 3. Inter. Pro 2 GO • Script that converts Inter. Pro IDs into their corresponding GO IDs • Available at geneontology. org

Comments 4. Adding GO using Blast: • Need to identify related species that have experimental GO • Search database of experimental GO (should not transfer annotations with IEA, ISS, etc evidence codes) • Use a test set of sequences to identify Blast parameters (e. g. Evalues, expect, etc. ) for the full dataset 5. Combining GO from Inter. Pro. Scan & Blast: • Remove any duplicate annotations derived from Inter. Pro. Scan (IEA) and Blast (ISA). • Remove any “no data” (ND) annotations where you have added an annotation using Blast. Note: GO IEA annotations are continually updated (by manual review) and are considered out of date after one year.

For help with adding GO, contact Ag. Base.