Modeling Functional Genomics Datasets CVM 889 101 Lesson

- Slides: 20

Modeling Functional Genomics Datasets CVM 889 -101 Lesson 7 25 June 2007 Fiona Mc. Carthy

Lesson 7 Outline Functional genomics modeling I: a eukaryotic example

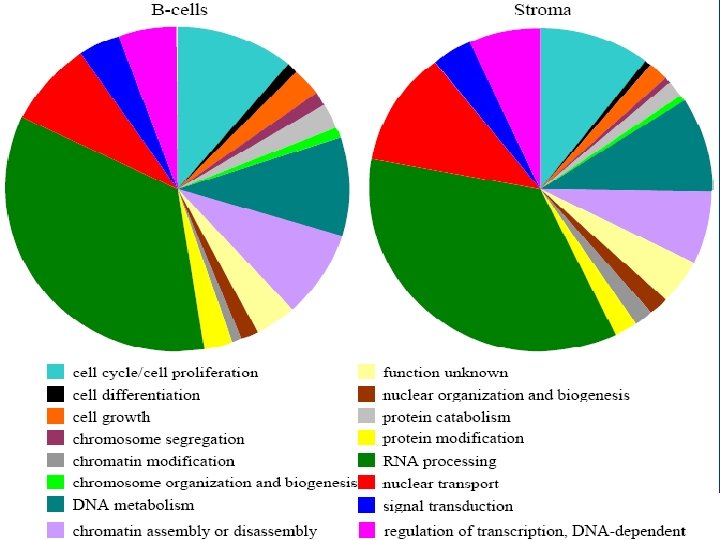
• identified 5, 198 proteins from chicken bursa – 1, 753 were B cell specific – 1, 972 were stroma specific – 1, 473 (28%) were shared between the two • identified 114 transcription factors (TFs) – 42 of the bursal B cell TFs have not been reported before in any B cells • modeled Programmed Cell Death (PCD), cell differentiation and proliferation, and transcriptional activation • confirmed the in vivo expression of 4, 006 “predicted”, and 6, 623 ab initio, ORFs

Introduction • cellular processes in complex eukaryotes do not occur in isolation, but are directed by soluble factors and interactions between cells. • understanding the biology of higher eukaryotes requires progressing to whole organ proteomics. • used differential detergent fractionationmultidimensional protein identification technology (DDF-Mud. PIT) to model the bursal proteome.

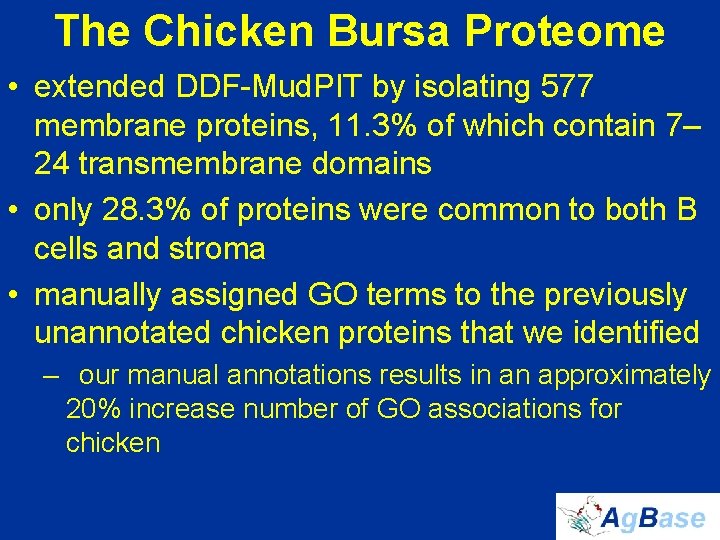
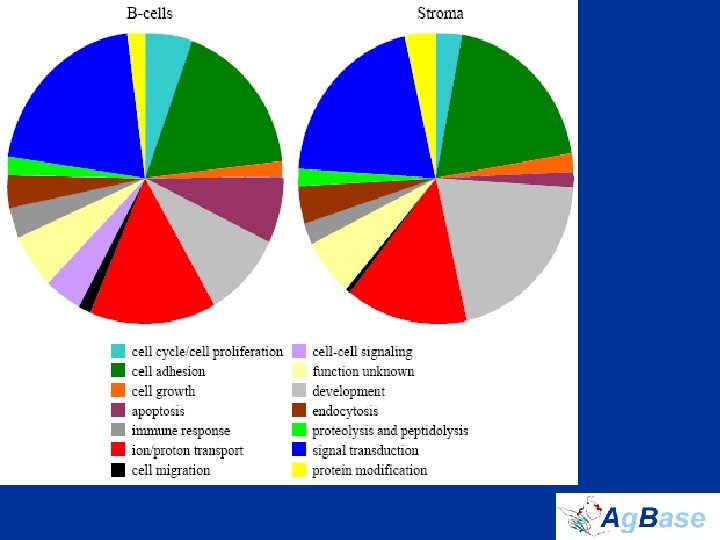
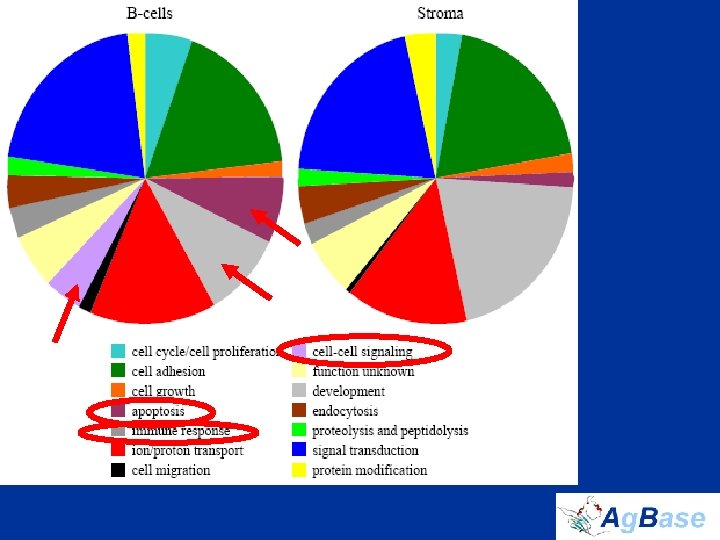
The Chicken Bursa Proteome • extended DDF-Mud. PIT by isolating 577 membrane proteins, 11. 3% of which contain 7– 24 transmembrane domains • only 28. 3% of proteins were common to both B cells and stroma • manually assigned GO terms to the previously unannotated chicken proteins that we identified – our manual annotations results in an approximately 20% increase number of GO associations for chicken

DDF-Mud. PIT: capturing the proteome

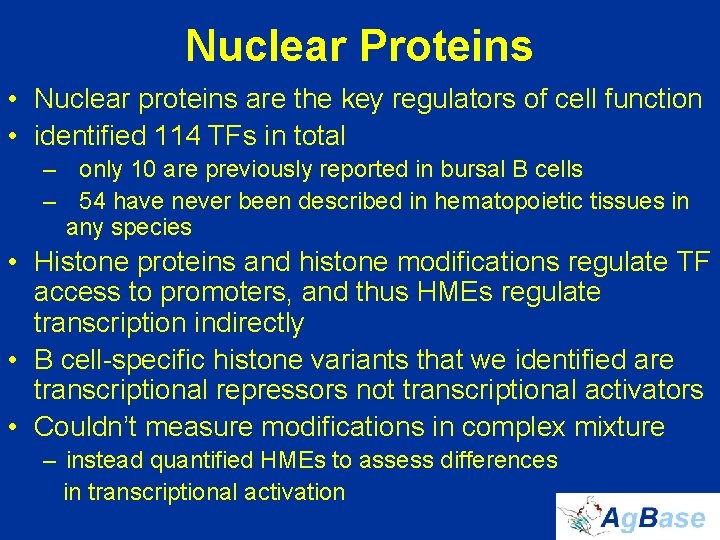
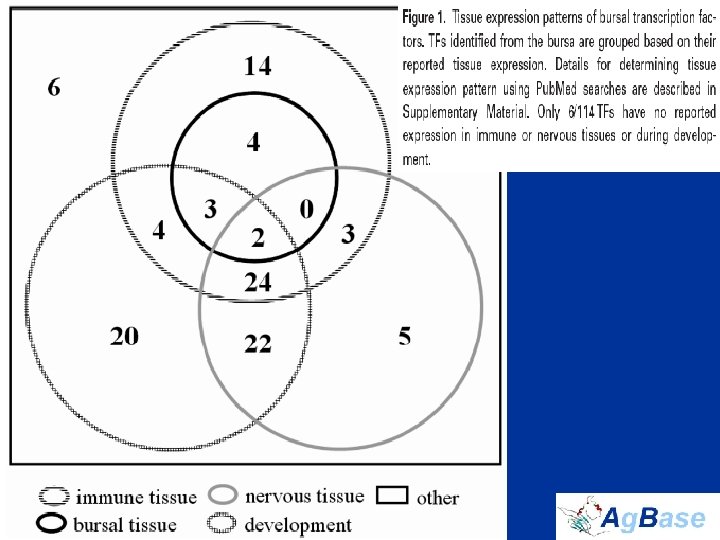
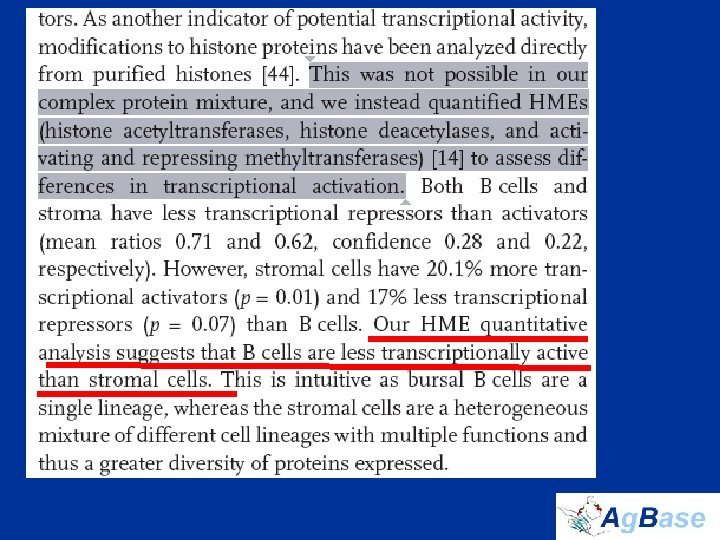
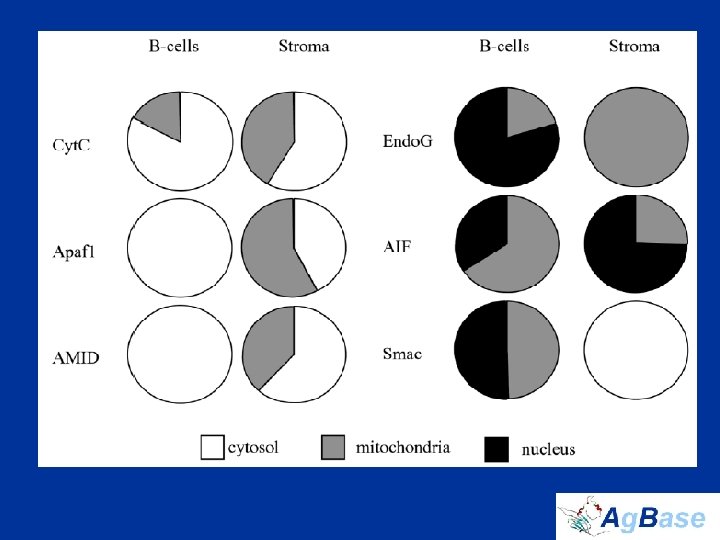
Nuclear Proteins • Nuclear proteins are the key regulators of cell function • identified 114 TFs in total – only 10 are previously reported in bursal B cells – 54 have never been described in hematopoietic tissues in any species • Histone proteins and histone modifications regulate TF access to promoters, and thus HMEs regulate transcription indirectly • B cell-specific histone variants that we identified are transcriptional repressors not transcriptional activators • Couldn’t measure modifications in complex mixture – instead quantified HMEs to assess differences in transcriptional activation


identified 114 TFs


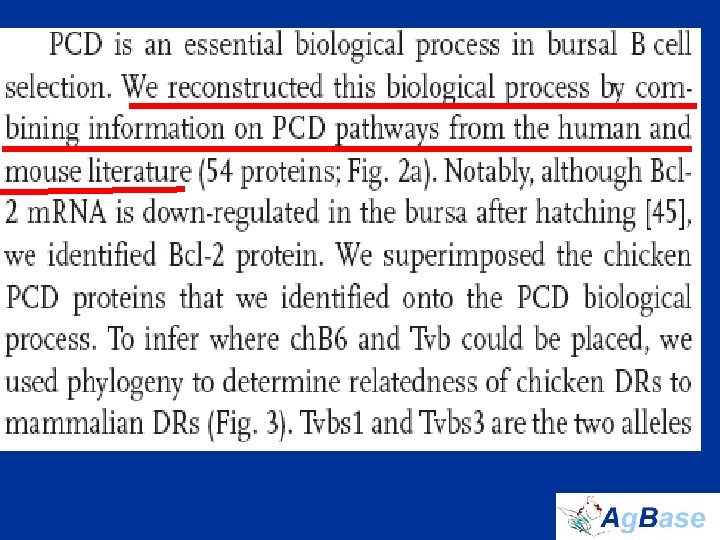
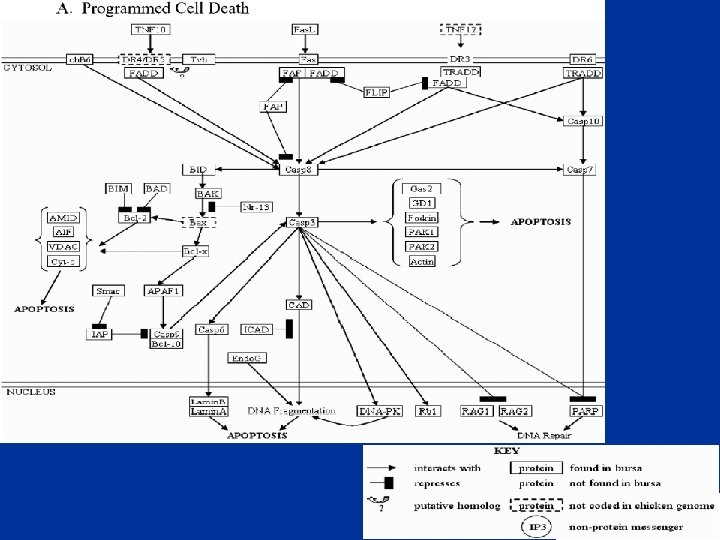
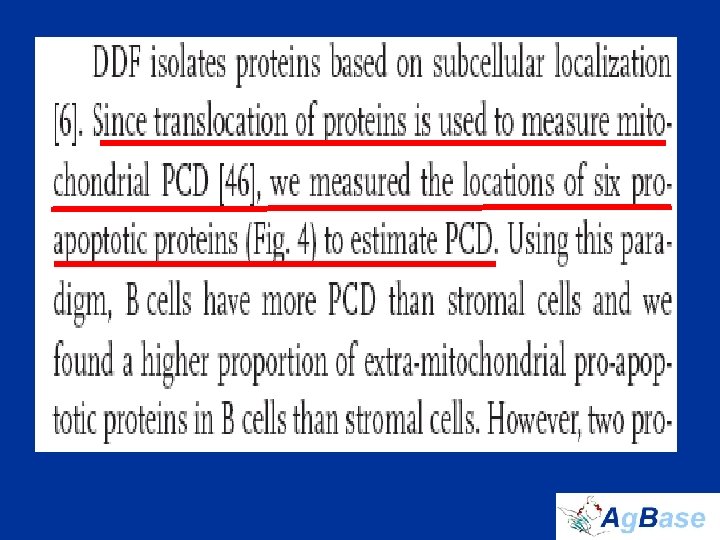
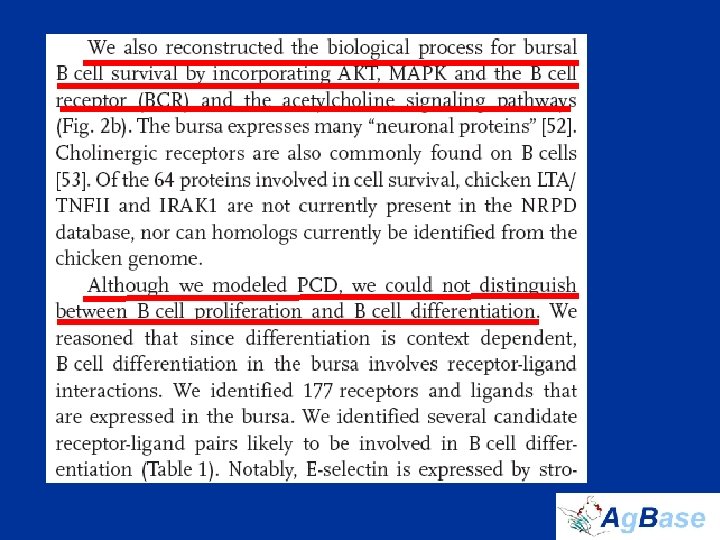
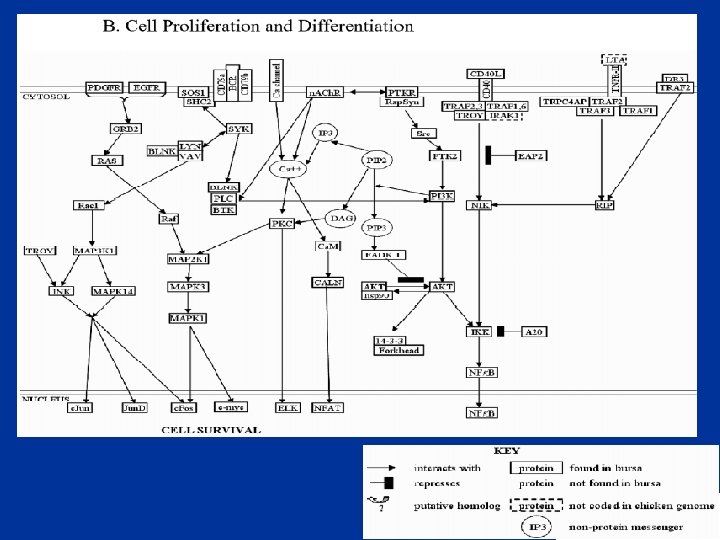
The Chicken Bursa • Site of B cell development and initial differentiation • Expect to see: – Programmed Cell Death (PCD) – Cell differentiation – Cell proliferation • biological process









Modeling Your Data • • What do you expect from your system? What do you see in your system? What is missing that you expect to see? What is there that you don’t expect to see?