Metagenome Data Analysis at the DOE JGI moving






- Slides: 20

Metagenome Data Analysis at the DOE JGI moving away from the streetlight Nikos Kyrpides Prokaryotic Program DOE Joint Genome Institute

Informatics Challenges Quality Integration Scaling Annotations Data Management # genes and genomes

Streetlight effect In 2005, John Ioannidis of the University of Ioannina in Greece examined the 45 most prominent studies published since 1990 in the top medical journals and found that about one-third of them were ultimately refuted. If one were to look at all medical studies, it would be more like two-thirds, he says. And for some kinds of leadingedge studies, like those linking a disease to a specific gene, wrongness infects 90 percent or more.

Overview 1. Current approaches 2. The Challenge 3. The JGI effort …and the possibilities

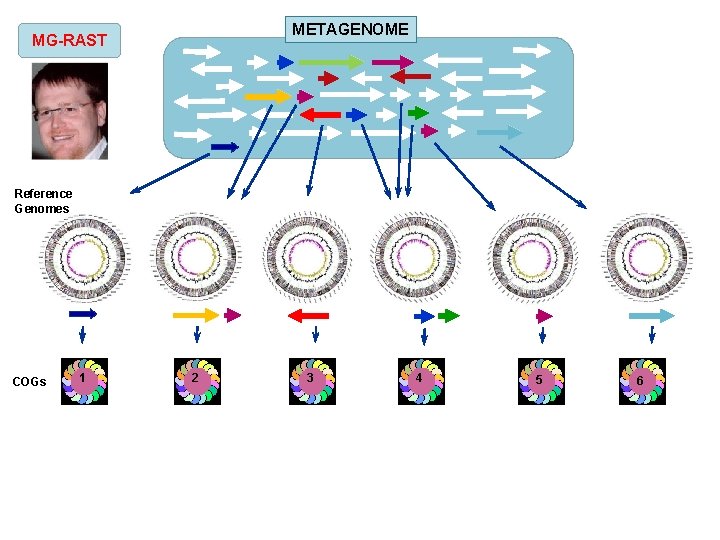
METAGENOME MG-RAST Reference Genomes COGs 1 2 3 4 5 6

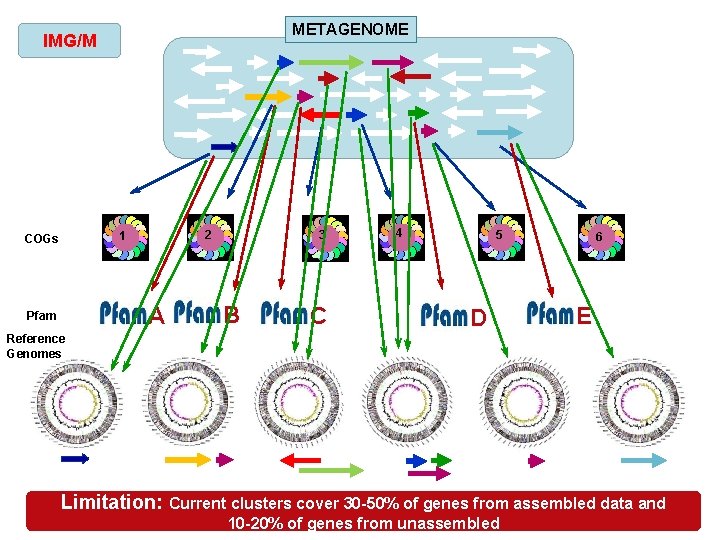
METAGENOME IMG/M 2 1 COGs A Pfam 3 B C 4 5 D 6 E Reference Genomes Limitation: Current clusters cover 30 -50% of genes from assembled data and 10 -20% of genes from unassembled

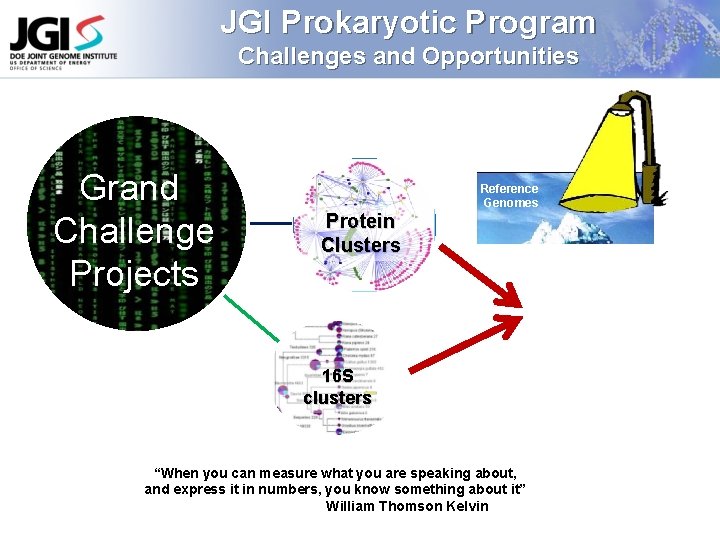
JGI Prokaryotic Program Challenges and Opportunities Grand Challenge Projects Reference Genomes Protein Clusters 16 S clusters “When you can measure what you are speaking about, and express it in numbers, you know something about it” William Thomson Kelvin

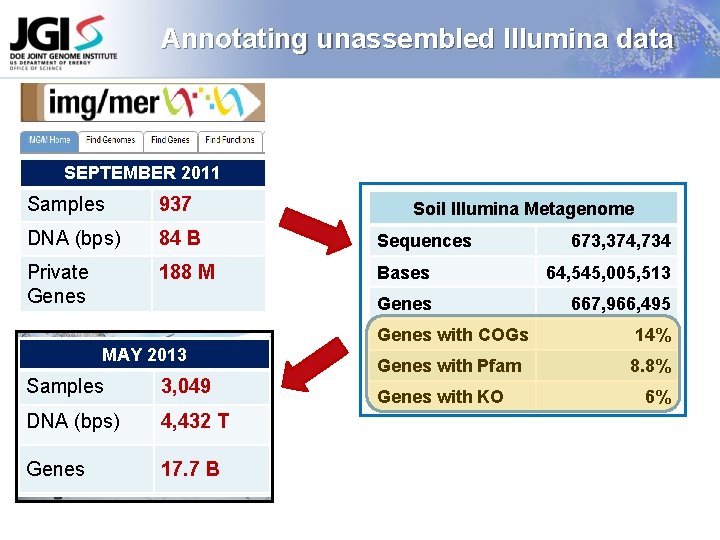
Annotating unassembled Illumina data SEPTEMBER 2011 Samples 937 DNA (bps) 84 B Sequences Private Genes 188 M Bases 64, 545, 005, 513 Genes 667, 966, 495 SEPTEMBER MAY 20132012 Samples 2, 112 3, 049 DNA (bps) 1, 266 4, 432 T Genes 8. 2 B 17. 7 B Soil Illumina Metagenome 673, 374, 734 Genes with COGs 14% Genes with Pfam 8. 8% Genes with KO 6%

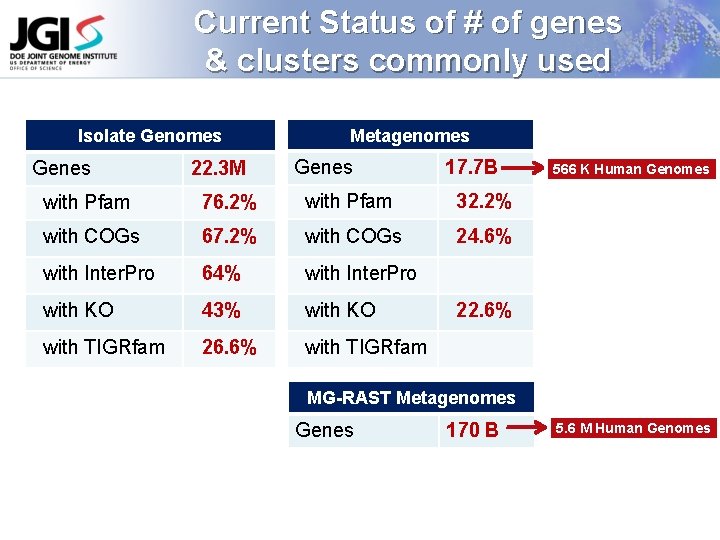
Current Status of # of genes & clusters commonly used Isolate Genomes Genes 22. 3 M Metagenomes Genes 17. 7 B with Pfam 76. 2% with Pfam 32. 2% with COGs 67. 2% with COGs 24. 6% with Inter. Pro 64% with Inter. Pro with KO 43% with KO with TIGRfam 26. 6% with TIGRfam 566 K Human Genomes 22. 6% MG-RAST Metagenomes Genes 170 B 5. 6 M Human Genomes

Where do we go from here?

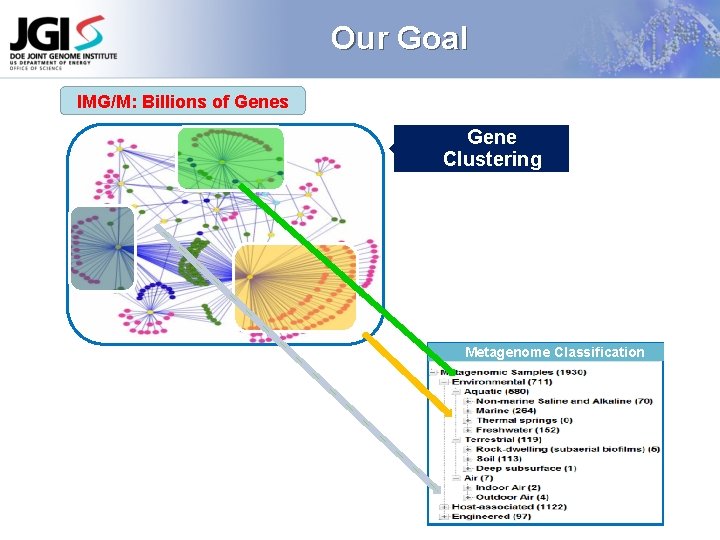
Our Goal IMG/M: Billions of Genes Gene Clustering Metagenome Classification

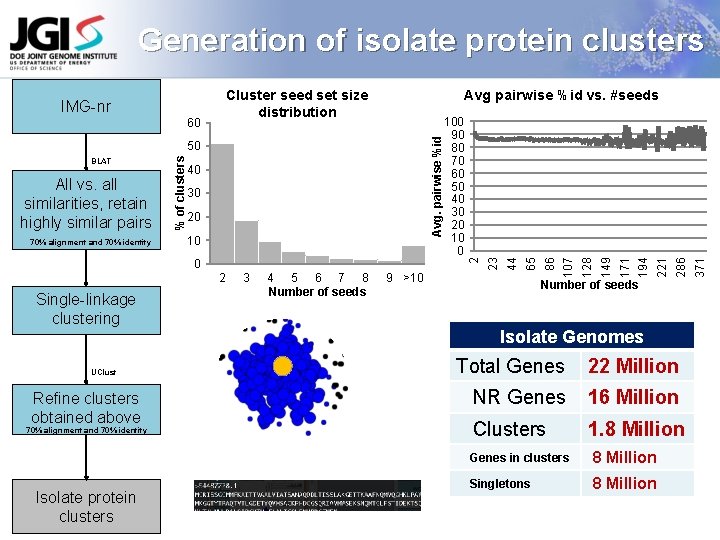
Generation of isolate protein clusters Cluster seed set size distribution Single-linkage clustering UClust Refine clusters obtained above 70% alignment and 70% identity Isolate protein clusters Number of seeds Isolate Genomes Total Genes 22 Million NR Genes 16 Million Clusters 1. 8 Million Genes in clusters 8 Million Singletons 8 Million 371 286 9 >10 221 4 5 6 7 8 Number of seeds 194 3 171 2 149 0 128 10 107 20 86 30 65 70% alignment and 70% identity 40 44 All vs. all similarities, retain highly similar pairs % of clusters 50 BLAT 100 90 80 70 60 50 40 30 20 10 0 23 Avg. pairwise %id 60 Avg pairwise %id vs. #seeds 2 IMG-nr

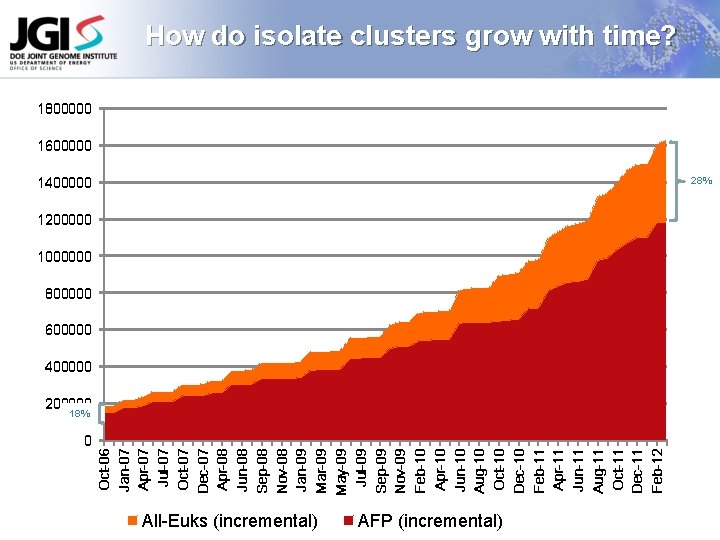
0 Oct-06 Jan-07 Apr-07 Jul-07 Oct-07 Dec-07 Apr-08 Jun-08 Sep-08 Nov-08 Jan-09 Mar-09 May-09 Jul-09 Sep-09 Nov-09 Feb-10 Apr-10 Jun-10 Aug-10 Oct-10 Dec-10 Feb-11 Apr-11 Jun-11 Aug-11 Oct-11 Dec-11 Feb-12 How do isolate clusters grow with time? 1800000 1600000 1400000 28% 1200000 1000000 800000 600000 400000 200000 18% All-Euks (incremental) AFP (incremental)

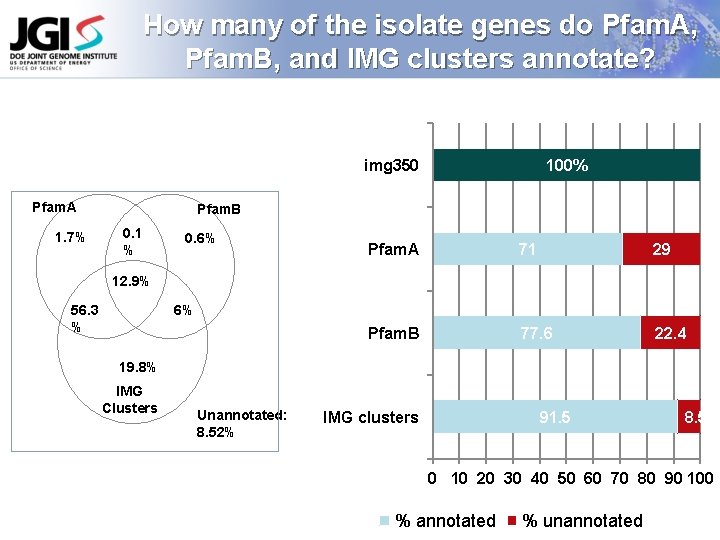
How many of the isolate genes do Pfam. A, Pfam. B, and IMG clusters annotate? img 350 Pfam. A 1. 7% 100% Pfam. B 0. 1 % 0. 6% Pfam. A 71 29 Pfam. B 77. 6 22. 4 12. 9% 56. 3 % 6% 19. 8% IMG Clusters Unannotated: 8. 52% IMG clusters 91. 5 8. 5 0 10 20 30 40 50 60 70 80 90 100 % annotated % unannotated

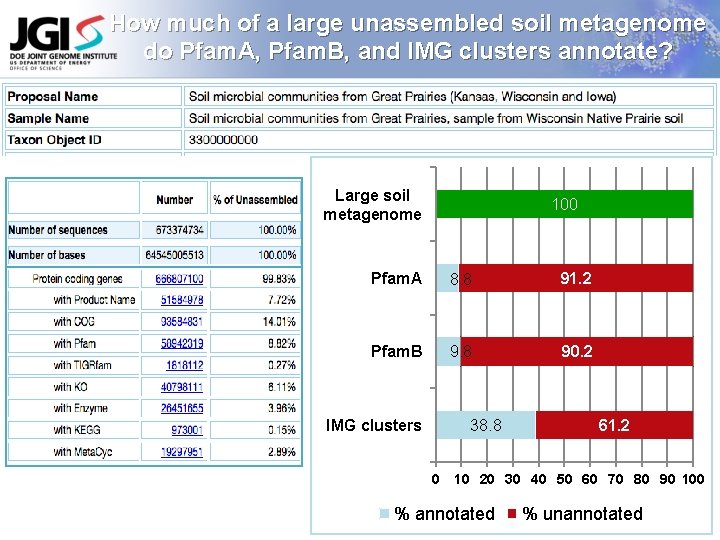
How much of a large unassembled soil metagenome do Pfam. A, Pfam. B, and IMG clusters annotate? Large soil metagenome 100 Pfam. A 8. 8 91. 2 Pfam. B 9. 8 90. 2 IMG clusters 38. 8 0 61. 2 10 20 30 40 50 60 70 80 90 100 % annotated % unannotated

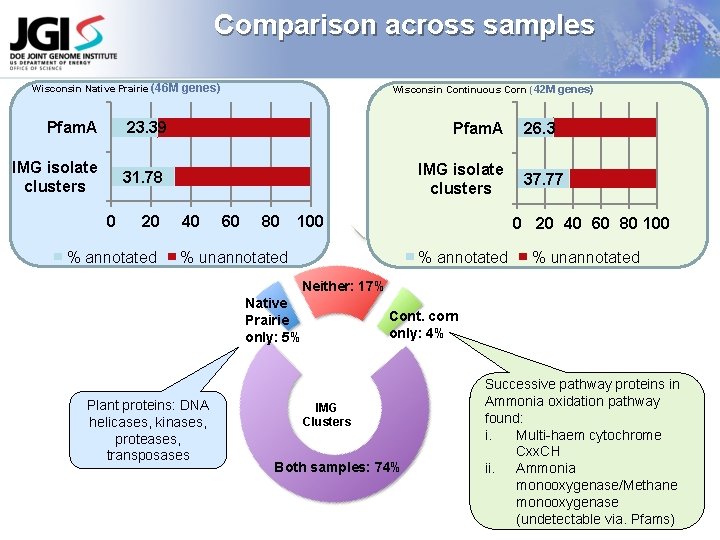
Comparison across samples Wisconsin Native Prairie (46 M genes) Wisconsin Continuous Corn (42 M genes) Pfam. A 23. 39 Pfam. A IMG isolate clusters 31. 78 IMG isolate clusters 0 20 % annotated 40 60 80 100 26. 3 37. 77 0 20 40 60 80 100 % unannotated Neither: 17% Native Prairie only: 5% Plant proteins: DNA helicases, kinases, proteases, transposases Cont. corn only: 4% IMG Clusters Both samples: 74% Successive pathway proteins in Ammonia oxidation pathway found: i. Multi-haem cytochrome Cxx. CH ii. Ammonia monooxygenase/Methane monooxygenase (undetectable via. Pfams)

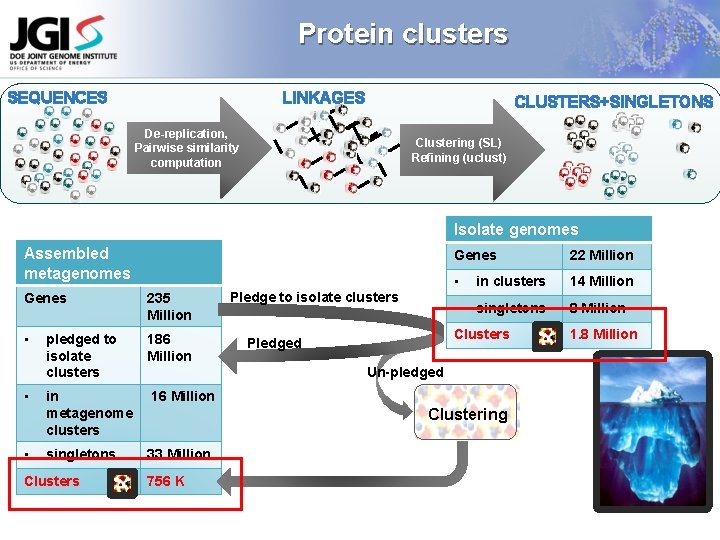
Next step Create new clusters from metagenomic genes that don’t have any hits to gene clusters from isolates Pledge all metagenomic genes to the clusters of isolates Create new clusters from the unpledged metagenomic genes

Protein clusters SEQUENCES LINKAGES CLUSTERS+SINGLETONS De-replication, Pairwise similarity computation Clustering (SL) Refining (uclust) Isolate genomes Assembled metagenomes Genes 235 Million • pledged to isolate clusters 186 Million in metagenome clusters 16 Million singletons 33 Million • • Clusters Pledge to isolate clusters Genes 22 Million • in clusters 14 Million • singletons 8 Million Clusters Pledged Un-pledged Clustering 756 K 1. 8 Million

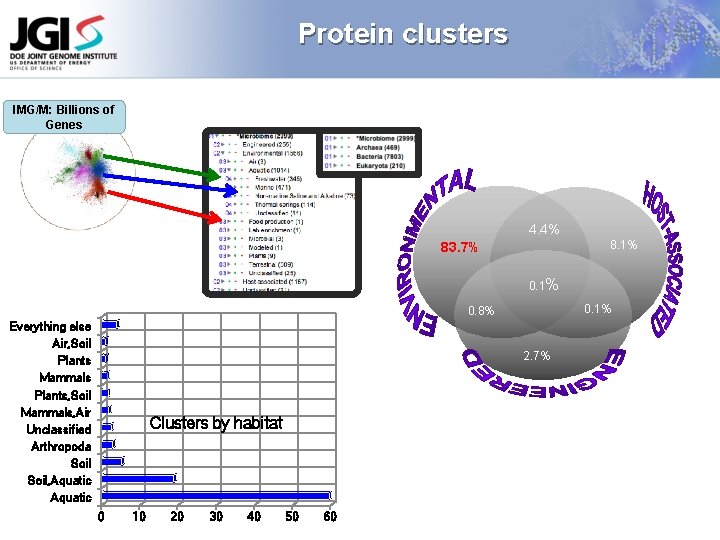
Protein clusters IMG/M: Billions of Genes 4. 4% 8. 1% 83. 7% 0. 1% 0. 8% Everything else Air, Soil Plants Mammals Plants, Soil Mammals, Air Unclassified Arthropoda Soil, Aquatic 2. 7% Clusters by habitat 0 10 20 30 40 50 60

Thank you