Meccanismi di Regolazione dellespressione genica Fase Nucleare Scelta

































- Slides: 58


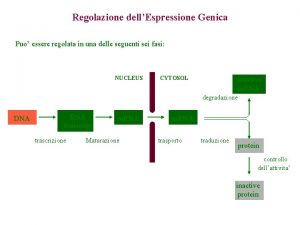
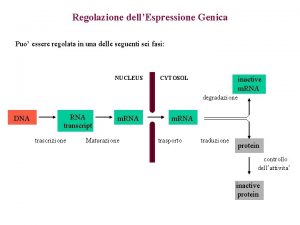
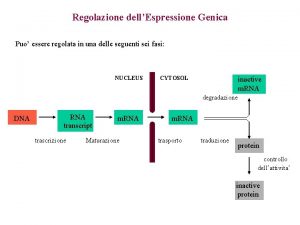
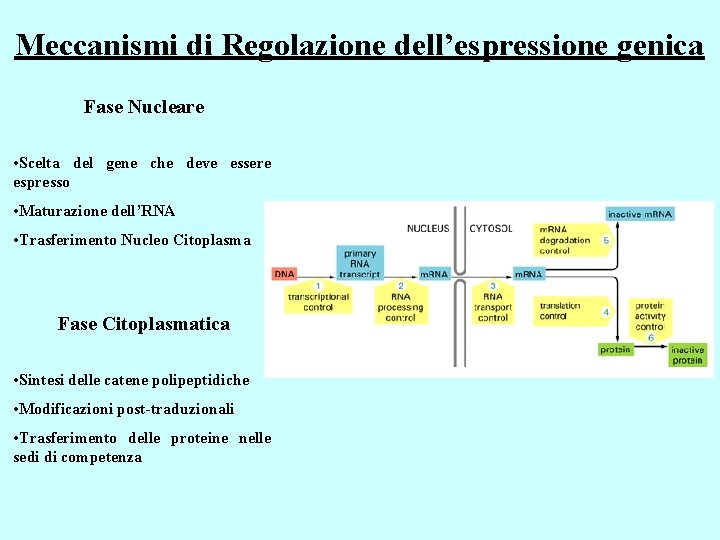
Meccanismi di Regolazione dell’espressione genica Fase Nucleare • Scelta del gene che deve essere espresso • Maturazione dell’RNA • Trasferimento Nucleo Citoplasma Fase Citoplasmatica • Sintesi delle catene polipeptidiche • Modificazioni post-traduzionali • Trasferimento delle proteine nelle sedi di competenza


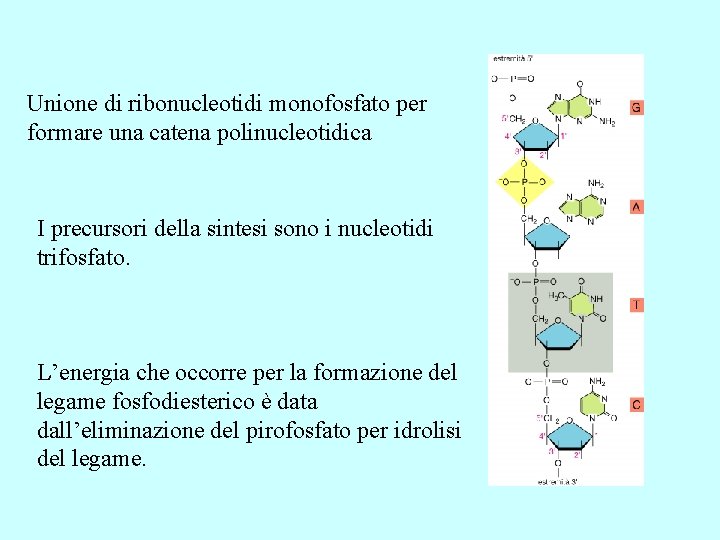
Unione di ribonucleotidi monofosfato per formare una catena polinucleotidica I precursori della sintesi sono i nucleotidi trifosfato. L’energia che occorre per la formazione del legame fosfodiesterico è data dall’eliminazione del pirofosfato per idrolisi del legame.

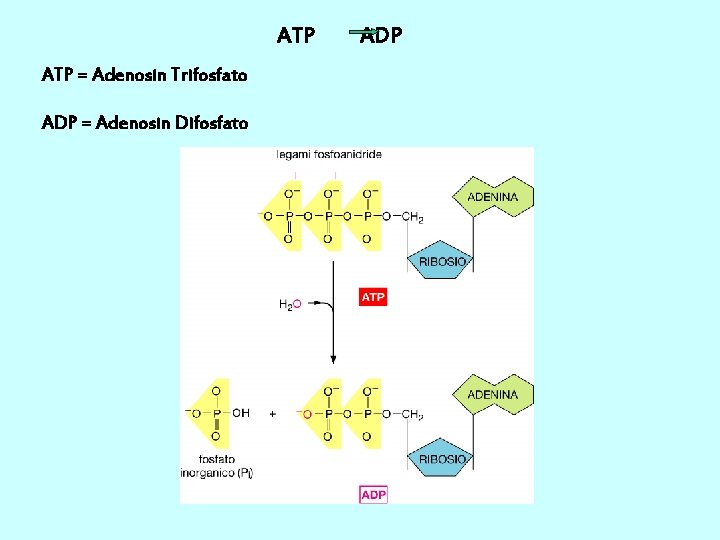
ATP = Adenosin Trifosfato ADP = Adenosin Difosfato ADP


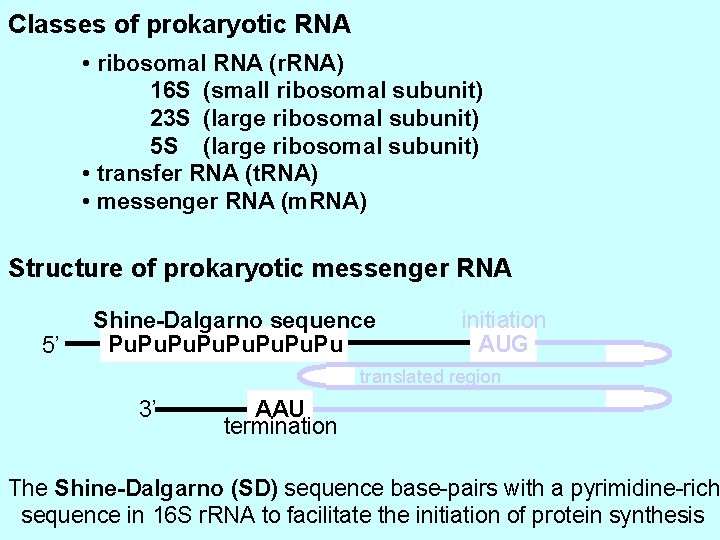
Classes of prokaryotic RNA • ribosomal RNA (r. RNA) 16 S (small ribosomal subunit) 23 S (large ribosomal subunit) 5 S (large ribosomal subunit) • transfer RNA (t. RNA) • messenger RNA (m. RNA) Structure of prokaryotic messenger RNA 5’ Shine-Dalgarno sequence Pu. Pu. Pu initiation AUG translated region 3’ AAU termination The Shine-Dalgarno (SD) sequence base-pairs with a pyrimidine-rich sequence in 16 S r. RNA to facilitate the initiation of protein synthesis

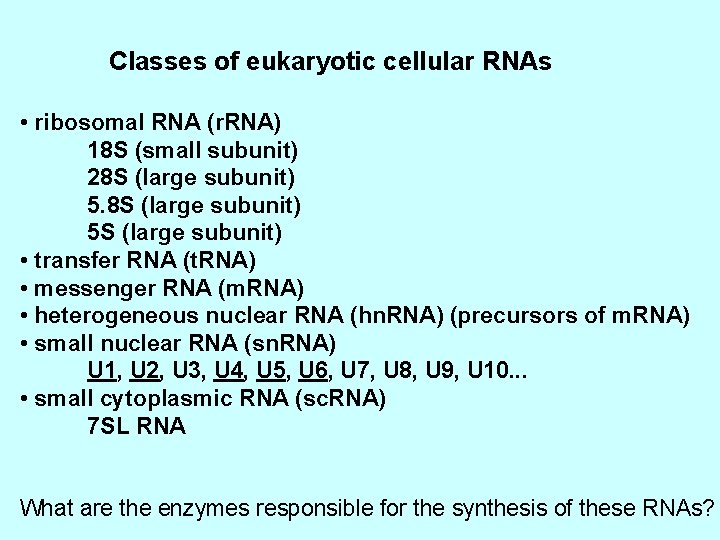
Classes of eukaryotic cellular RNAs • ribosomal RNA (r. RNA) 18 S (small subunit) 28 S (large subunit) 5 S (large subunit) • transfer RNA (t. RNA) • messenger RNA (m. RNA) • heterogeneous nuclear RNA (hn. RNA) (precursors of m. RNA) • small nuclear RNA (sn. RNA) U 1, U 2, U 3, U 4, U 5, U 6, U 7, U 8, U 9, U 10. . . • small cytoplasmic RNA (sc. RNA) 7 SL RNA What are the enzymes responsible for the synthesis of these RNAs?

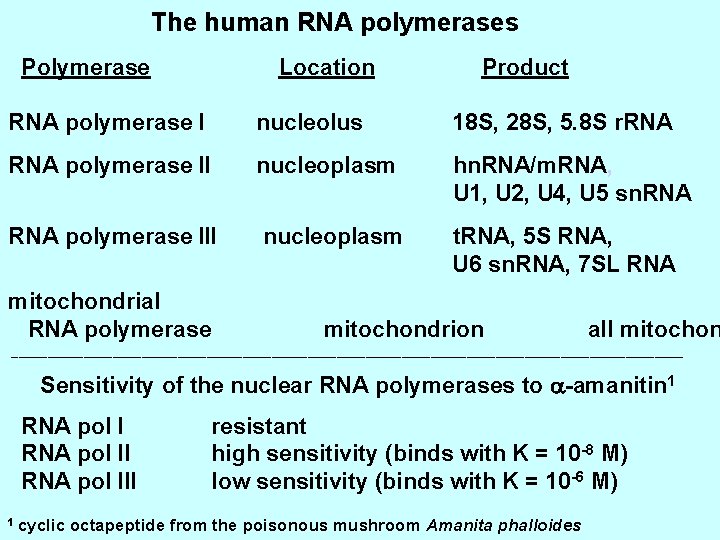
The human RNA polymerases Polymerase Location Product RNA polymerase I nucleolus 18 S, 28 S, 5. 8 S r. RNA polymerase II nucleoplasm hn. RNA/m. RNA, U 1, U 2, U 4, U 5 sn. RNA polymerase III nucleoplasm t. RNA, 5 S RNA, U 6 sn. RNA, 7 SL RNA mitochondrial RNA polymerase mitochondrion all mitochon _______________________________________________ Sensitivity of the nuclear RNA polymerases to a-amanitin 1 RNA pol III 1 cyclic resistant high sensitivity (binds with K = 10 -8 M) low sensitivity (binds with K = 10 -6 M) octapeptide from the poisonous mushroom Amanita phalloides

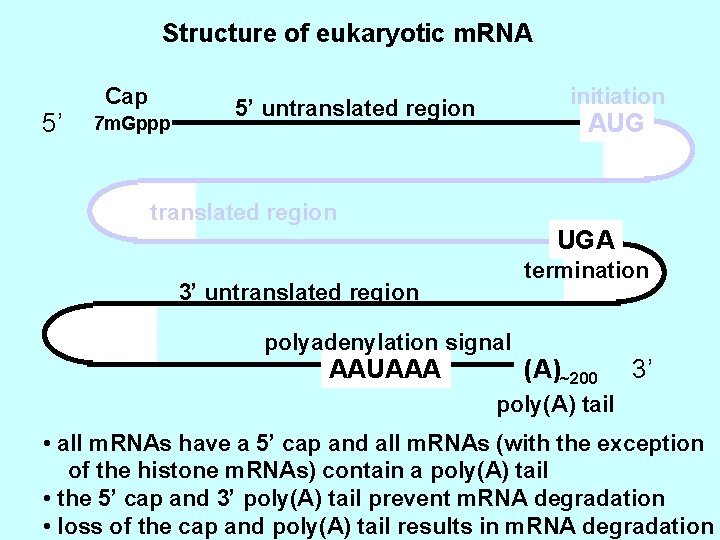
Structure of eukaryotic m. RNA 5’ Cap 7 m. Gppp initiation 5’ untranslated region AUG translated region UGA termination 3’ untranslated region polyadenylation signal AAUAAA (A)~200 3’ poly(A) tail • all m. RNAs have a 5’ cap and all m. RNAs (with the exception of the histone m. RNAs) contain a poly(A) tail • the 5’ cap and 3’ poly(A) tail prevent m. RNA degradation • loss of the cap and poly(A) tail results in m. RNA degradation




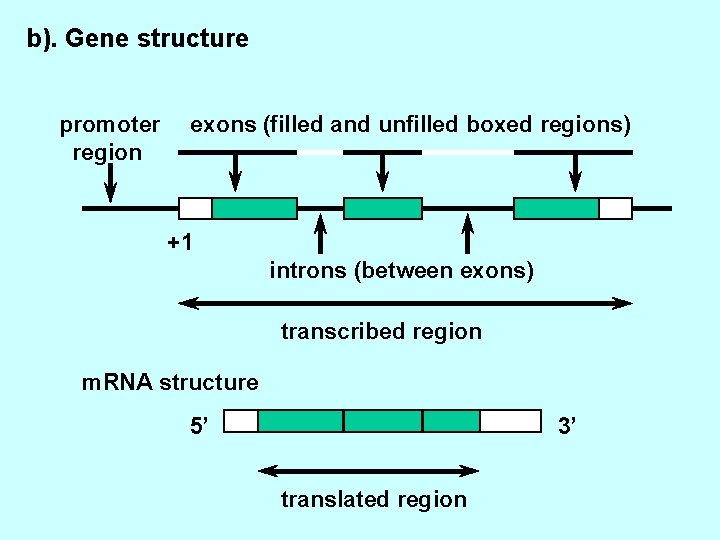
b). Gene structure promoter region exons (filled and unfilled boxed regions) +1 introns (between exons) transcribed region m. RNA structure 5’ 3’ translated region

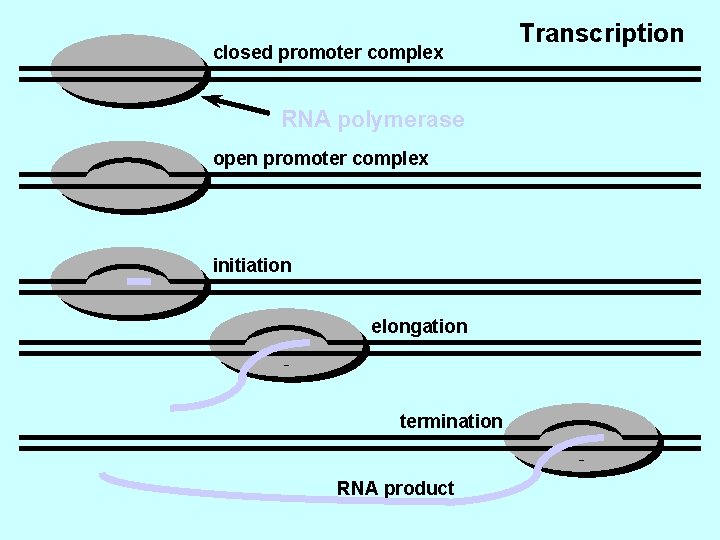
Legame al promotore della RNA polimerasi Apertura della doppia elica Inizio della sintesi Allungamento Terminazione

closed promoter complex RNA polymerase open promoter complex initiation elongation termination RNA product Transcription

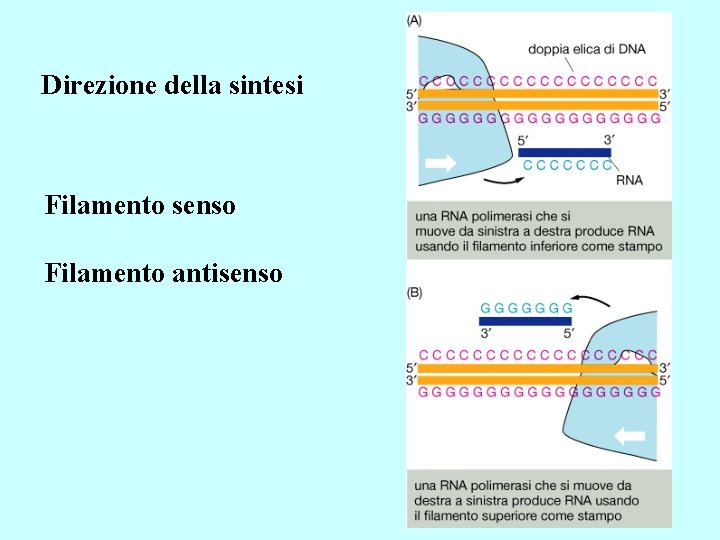
Direzione della sintesi Filamento senso Filamento antisenso


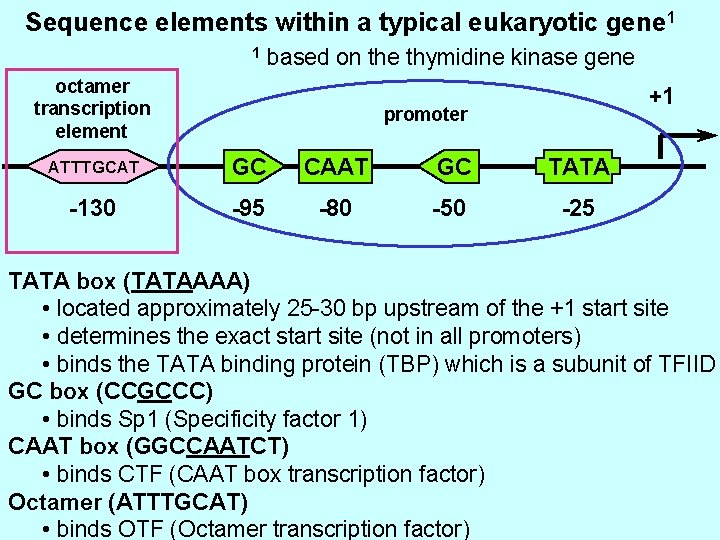
Sequence elements within a typical eukaryotic gene 1 1 based on the thymidine kinase gene octamer transcription element +1 promoter ATTTGCAT GC CAAT GC TATA -130 -95 -80 -50 -25 TATA box (TATAAAA) • located approximately 25 -30 bp upstream of the +1 start site • determines the exact start site (not in all promoters) • binds the TATA binding protein (TBP) which is a subunit of TFIID GC box (CCGCCC) • binds Sp 1 (Specificity factor 1) CAAT box (GGCCAATCT) • binds CTF (CAAT box transcription factor) Octamer (ATTTGCAT) • binds OTF (Octamer transcription factor)

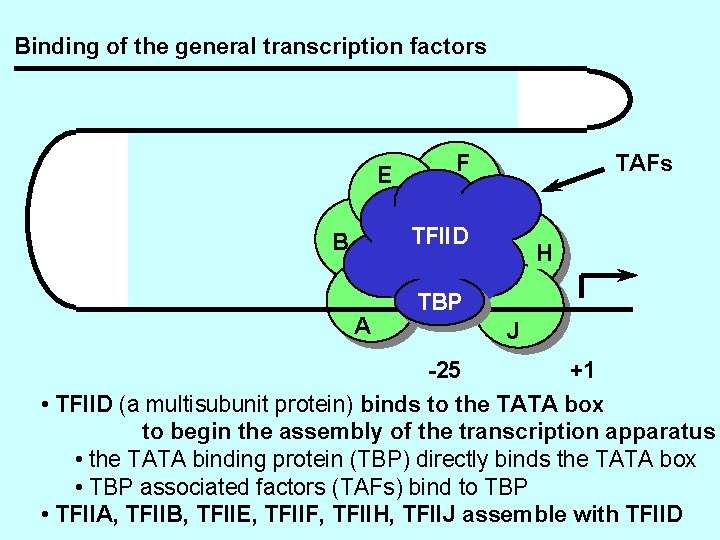
Proteins regulating eukaryotic m. RNA synthesis General transcription factors • TFIID (a multisubunit protein) binds to the TATA box to begin the assembly of the transcription apparatus • the TATA binding protein (TBP) directly binds the TATA box • TBP associated factors (TAFs) bind to TBP • TFIIA, TFIIB, TFIIE, TFIIF, TFIIH 1, TFIIJ assemble with TFIID RNA polymerase II binds the promoter region via the TFII’s Transcription factors binding to other promoter elements and transcription elements interact with proteins at the promoter and further stabilize (or inhibit) formation of a functional preinitiation complex 1 TFIIH is also involved in phosphorylation of RNA polymerase II, DNA repair (Cockayne syndrome mutations), and cell cycle regulation

Nome TF 2 D TBP + TAF Alias Chromosoma TAF 1 250 Xq 13. 1 TAF 1 L 250 like 9 p 21. 1 TAF 2 150 8 q 24. 12 TAF 3 140 10 p 15. 1 TAF 4 135 20 q 13. 33 TAF 4 B 105 18 q 11. 2 TAF 5 100 10 q 24 -10 q 25. 2 TAF 6 80 7 q 22. 1 TAF 6 L 11 q 12. 3 TAF 7 55 5 q 31 TAF 7 L 50 Xq 22. 1 TAF 8 43 6 p 21. 1 TAF 9 32 5 q 11. 2 -5 q 13. 1 TAF 10 30 11 p 15. 3 TAF 11 28 6 p 21. 31 TAF 12 20 -15 1 p 35. 3 TAF 13 18 1 p 13. 3 TAF 15 68 17 q 11. 1 -q 11. 2

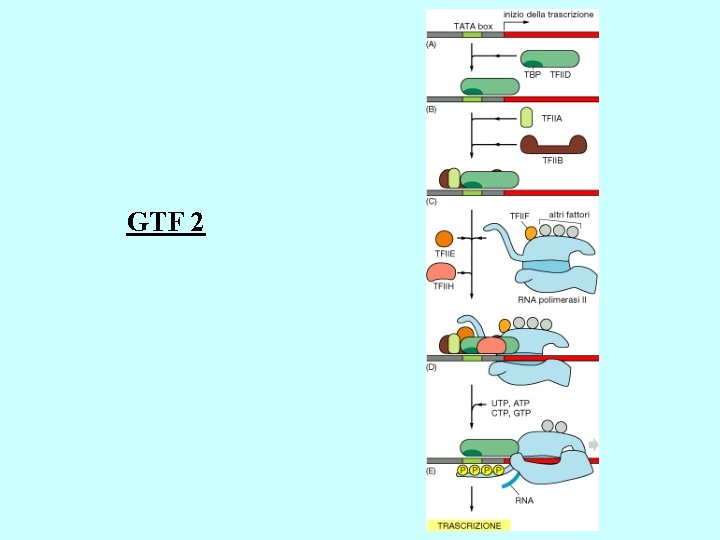
GTF 2


Binding of the general transcription factors E F TAFs TFIID B A H TBP J -25 +1 • TFIID (a multisubunit protein) binds to the TATA box to begin the assembly of the transcription apparatus • the TATA binding protein (TBP) directly binds the TATA box • TBP associated factors (TAFs) bind to TBP • TFIIA, TFIIB, TFIIE, TFIIF, TFIIH, TFIIJ assemble with TFIID

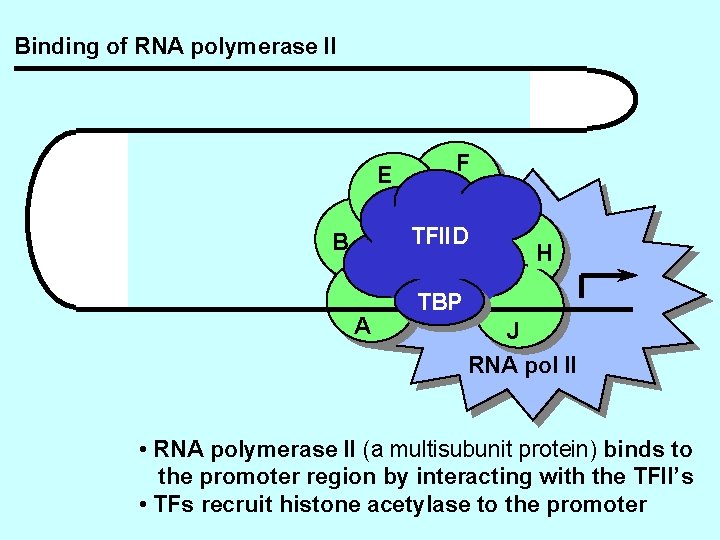
Binding of RNA polymerase II E F TFIID B A H TBP J RNA pol II • RNA polymerase II (a multisubunit protein) binds to the promoter region by interacting with the TFII’s • TFs recruit histone acetylase to the promoter


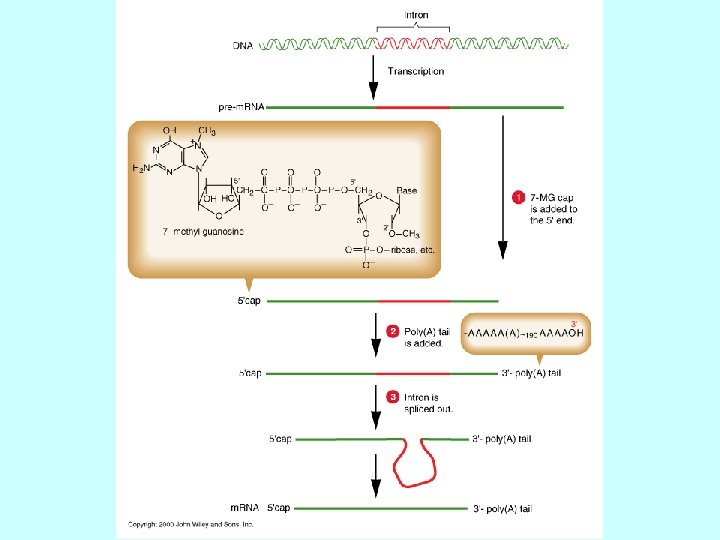
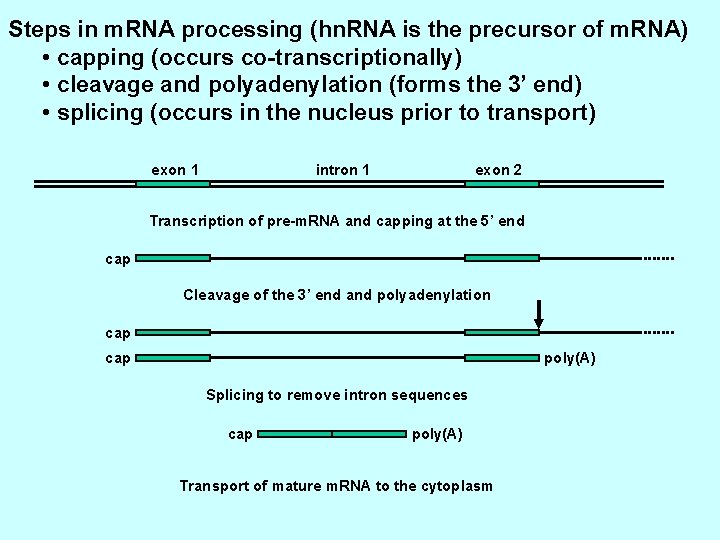
Steps in m. RNA processing (hn. RNA is the precursor of m. RNA) • capping (occurs co-transcriptionally) • cleavage and polyadenylation (forms the 3’ end) • splicing (occurs in the nucleus prior to transport) exon 1 intron 1 exon 2 Transcription of pre-m. RNA and capping at the 5’ end cap Cleavage of the 3’ end and polyadenylation cap poly(A) Splicing to remove intron sequences cap poly(A) Transport of mature m. RNA to the cytoplasm

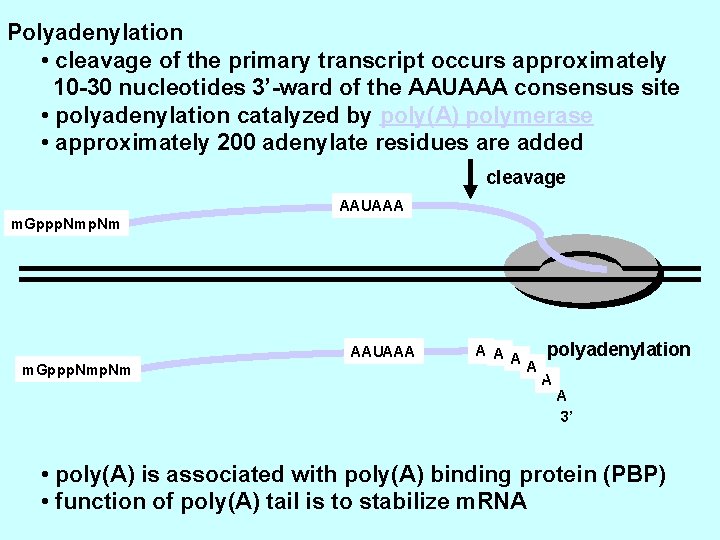
Polyadenylation • cleavage of the primary transcript occurs approximately 10 -30 nucleotides 3’-ward of the AAUAAA consensus site • polyadenylation catalyzed by poly(A) polymerase • approximately 200 adenylate residues are added cleavage AAUAAA m. Gppp. Nmp. Nm A A polyadenylation A A 3’ • poly(A) is associated with poly(A) binding protein (PBP) • function of poly(A) tail is to stabilize m. RNA


Splicing Rimozione di un introne attraverso due reazioni sequenziali di trasferimento di fosfato, note come transesterificazioni. Queste uniscono due esoni rimuovendo l’introne come un “cappio”




GENE PREDICTION Gene. Scan Grail. EXP Promoter 2. 0 Omiga 2. 0 Gene. Mark F GENE SH




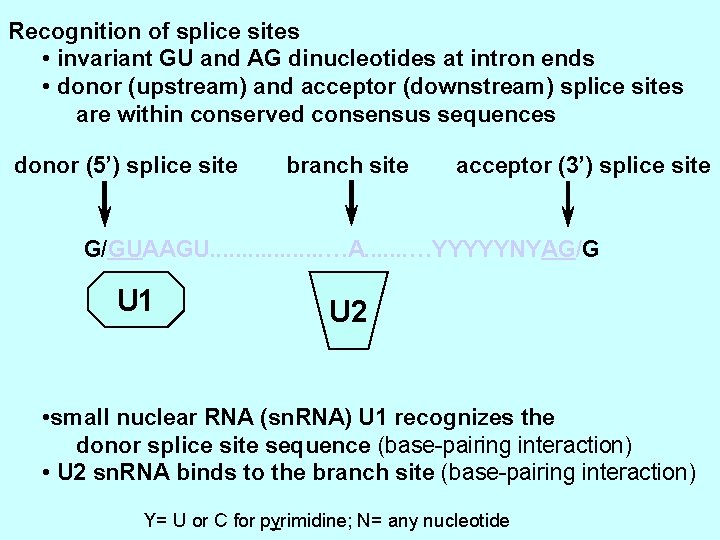
Recognition of splice sites • invariant GU and AG dinucleotides at intron ends • donor (upstream) and acceptor (downstream) splice sites are within conserved consensus sequences donor (5’) splice site branch site acceptor (3’) splice site G/GUAAGU. . . . …A. . . . …YYYYYNYAG/G U 1 U 2 • small nuclear RNA (sn. RNA) U 1 recognizes the donor splice site sequence (base-pairing interaction) • U 2 sn. RNA binds to the branch site (base-pairing interaction) Y= U or C for pyrimidine; N= any nucleotide

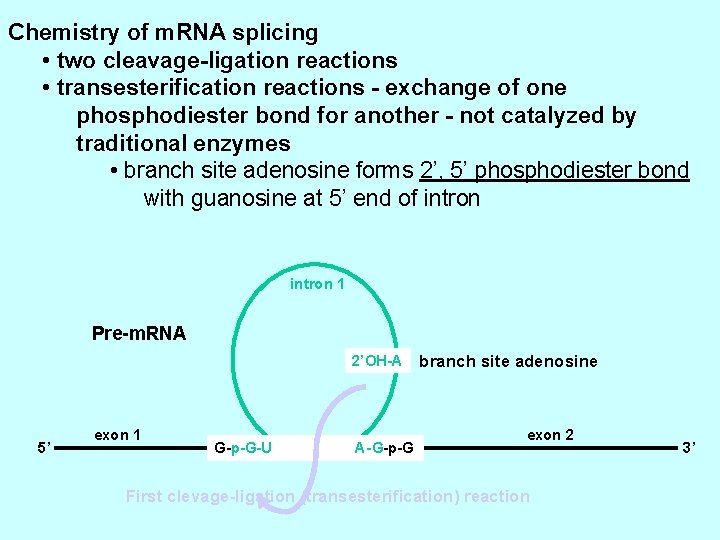
Chemistry of m. RNA splicing • two cleavage-ligation reactions • transesterification reactions - exchange of one phosphodiester bond for another - not catalyzed by traditional enzymes • branch site adenosine forms 2’, 5’ phosphodiester bond with guanosine at 5’ end of intron 1 Pre-m. RNA 2’OH-A 5’ exon 1 G-p-G-U - A-G-p-G branch site adenosine exon 2 First clevage-ligation (transesterification) reaction 3’

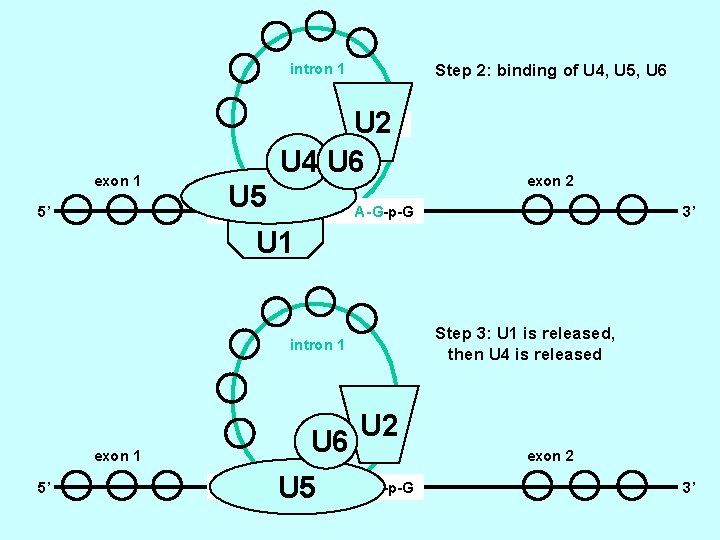
intron 1 Step 2: binding of U 4, U 5, U 6 U 2 U 4 U 6 2’OH-A exon 1 5’ U 5 G-p-G-U - exon 2 A-G-p-G 3’ U 1 Step 3: U 1 is released, then U 4 is released intron 1 2’OH-A U 6 exon 1 5’ G-p-G-U - U 5 U 2 exon 2 A-G-p-G 3’

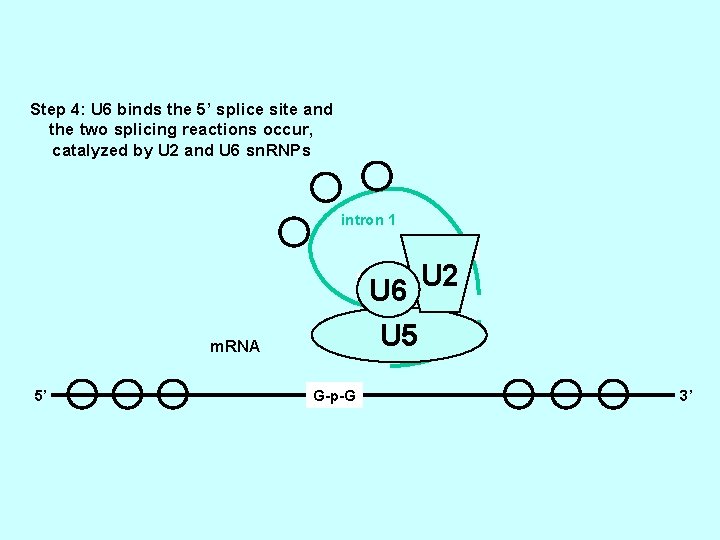
Step 4: U 6 binds the 5’ splice site and the two splicing reactions occur, catalyzed by U 2 and U 6 sn. RNPs intron 1 2’OH-A U 2 U 6 U-G-5’-p-2’-A A m. RNA 5’ 3’ G-A G-p-G U 5 3’

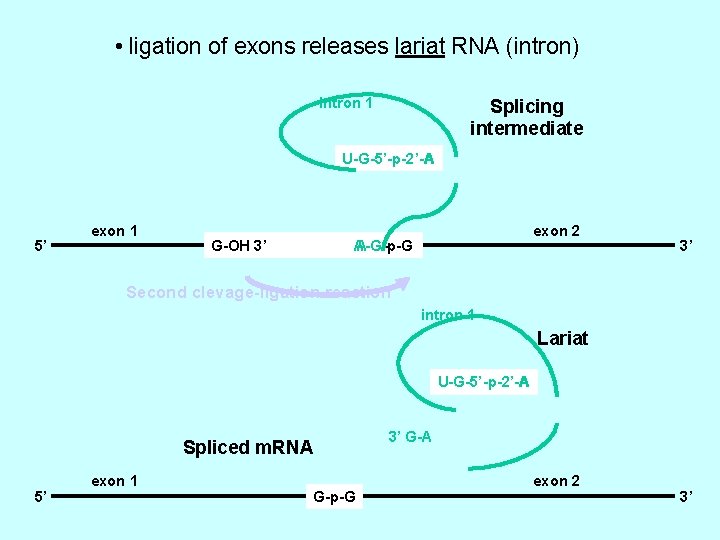
• ligation of exons releases lariat RNA (intron) intron 1 Splicing intermediate U-G-5’-p-2’-A A 5’ exon 1 G-OH O 3’ exon 2 A-G-p-G A - 3’ Second clevage-ligation reaction intron 1 Lariat U-G-5’-p-2’-A A Spliced m. RNA 5’ exon 1 G-p-G 3’ G-A exon 2 3’


Trans-splicing Nei protozoi e in un nematode






Differenti molecole di m. RNA dallo stesso gene Splicing alternativo Uso di promotori alternativi Uso di segnali di poliadenilazione alternativi





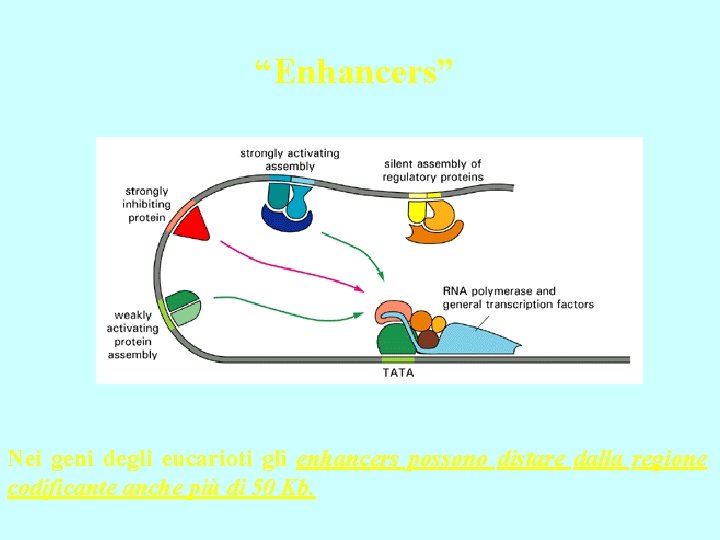
“Enhancers” Nei geni degli eucarioti gli enhancers possono distare dalla regione codificante anche più di 50 Kb.



 Operone lac è trp zanichelli
Operone lac è trp zanichelli Regolazione espressione genica eucarioti
Regolazione espressione genica eucarioti Regolazione genica nei procarioti
Regolazione genica nei procarioti Test di appercezione tematica tavole
Test di appercezione tematica tavole La nutrizione
La nutrizione Meccanismi epigenetici
Meccanismi epigenetici Valvola di von gubaroff
Valvola di von gubaroff Meccanismi locali di ribaltamento
Meccanismi locali di ribaltamento Modello mintzberg
Modello mintzberg Regolazione omeometrica
Regolazione omeometrica Teoria bifattoriale della regolazione delle emozioni
Teoria bifattoriale della regolazione delle emozioni La regolazione degli affetti e la riparazione del sé
La regolazione degli affetti e la riparazione del sé Tesabase
Tesabase Teoria della scelta razionale
Teoria della scelta razionale Studiare unife my desk
Studiare unife my desk Quesiti a risposta multipla
Quesiti a risposta multipla Stato patrimoniale
Stato patrimoniale Fase fase perkembangan antropologi
Fase fase perkembangan antropologi Fase fase proses kompilasi
Fase fase proses kompilasi Rumus beda fase
Rumus beda fase Contoh fase diam pada klt
Contoh fase diam pada klt Elemen proses pemecahan masalah
Elemen proses pemecahan masalah Leer y transcribir
Leer y transcribir Fase heurística y fase hermenéutica
Fase heurística y fase hermenéutica Fase fase pengambilan keputusan
Fase fase pengambilan keputusan Ebb phase adalah
Ebb phase adalah 3 fase naar 1 fase transformator
3 fase naar 1 fase transformator Dna mutazioni
Dna mutazioni Expresion genica
Expresion genica Uma ovelha gerada na inglaterra chamada tracy
Uma ovelha gerada na inglaterra chamada tracy Sexo
Sexo Duplicazione genica
Duplicazione genica Collo di bottiglia
Collo di bottiglia Compensacion de dosis genica
Compensacion de dosis genica Duplicazione genica
Duplicazione genica Iaiaou
Iaiaou Espressione genica
Espressione genica Expresión génica
Expresión génica Fusione nucleare
Fusione nucleare Frase nucleare scuola primaria
Frase nucleare scuola primaria Interazione nucleare debole
Interazione nucleare debole Centrale nucleare schema
Centrale nucleare schema Unità di misura in fisica nucleare
Unità di misura in fisica nucleare Funzionamento centrale nucleare
Funzionamento centrale nucleare Forza debole
Forza debole Fusione nucleare schema
Fusione nucleare schema Cosa fa il nucleo
Cosa fa il nucleo Interazione nucleare debole
Interazione nucleare debole Energia nucleare riassunto
Energia nucleare riassunto Detector de radiatii nucleare
Detector de radiatii nucleare Marcato pleomorfismo nucleare
Marcato pleomorfismo nucleare Ran gef and ran gap
Ran gef and ran gap Fissione nucleare
Fissione nucleare Interazione nucleare debole
Interazione nucleare debole Isotopi
Isotopi Fusione nucleare
Fusione nucleare Medicina nucleare civile brescia
Medicina nucleare civile brescia Fissione nucleare
Fissione nucleare Interazione nucleare debole
Interazione nucleare debole