Introduction to Protein Structure Rui Kuang Why do


- Slides: 17

Introduction to Protein Structure Rui Kuang

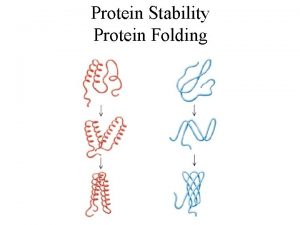
Why do we study protein structure • Protein – Derived from Greek word proteios meaning “of the first rank” in 1838 by Jöns J. Berzelius. • Crucial in all biological processes, such as Enzymatic catalysis, transport and storage, immune protection…… • Functions depend on structures --- structure can help us to understand function

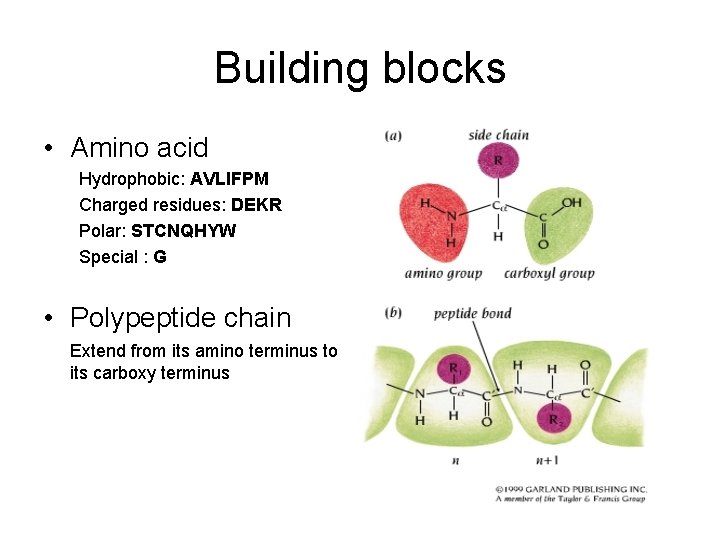
Building blocks • Amino acid Hydrophobic: AVLIFPM Charged residues: DEKR Polar: STCNQHYW Special : G • Polypeptide chain Extend from its amino terminus to its carboxy terminus

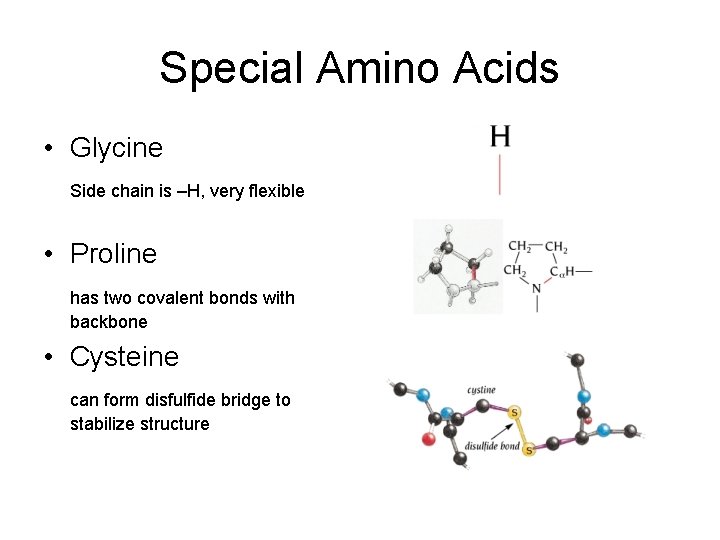
Special Amino Acids • Glycine Side chain is –H, very flexible • Proline has two covalent bonds with backbone • Cysteine can form disfulfide bridge to stabilize structure

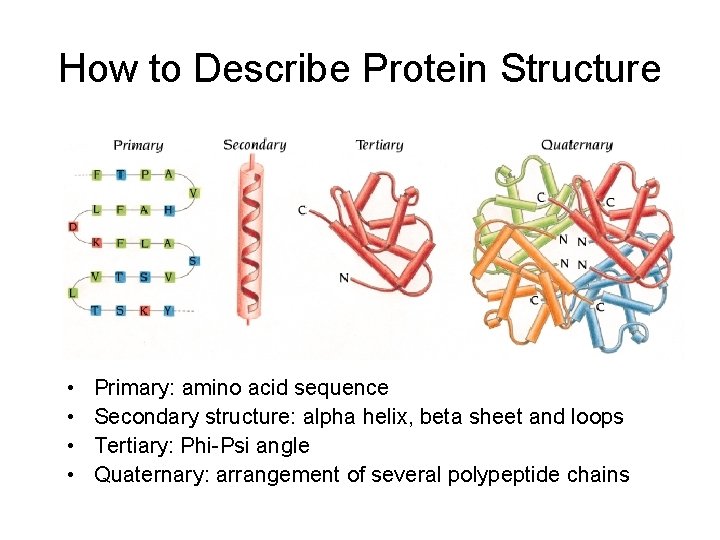
How to Describe Protein Structure • • Primary: amino acid sequence Secondary structure: alpha helix, beta sheet and loops Tertiary: Phi-Psi angle Quaternary: arrangement of several polypeptide chains

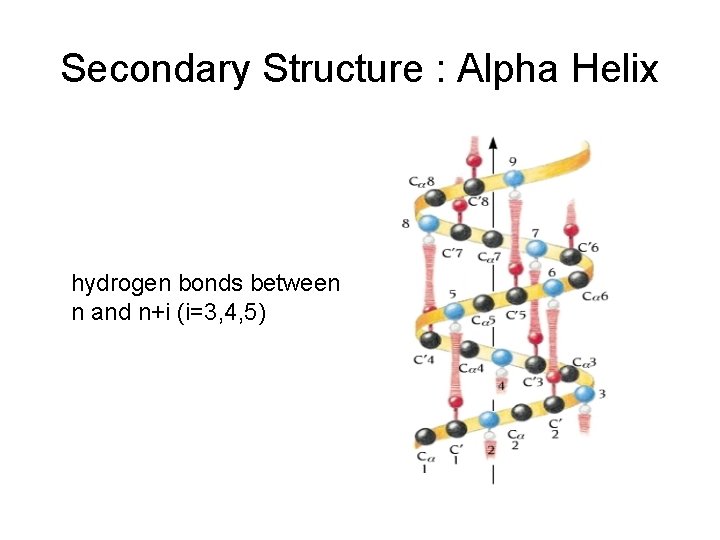
Secondary Structure : Alpha Helix hydrogen bonds between n and n+i (i=3, 4, 5)

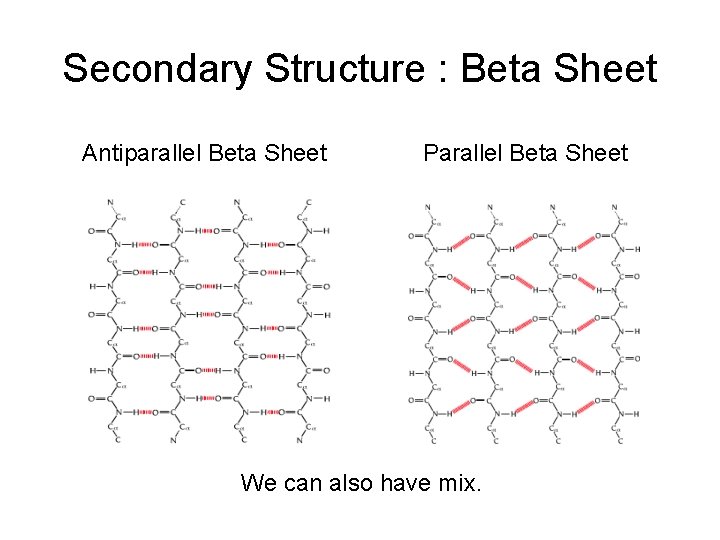
Secondary Structure : Beta Sheet Antiparallel Beta Sheet Parallel Beta Sheet We can also have mix.

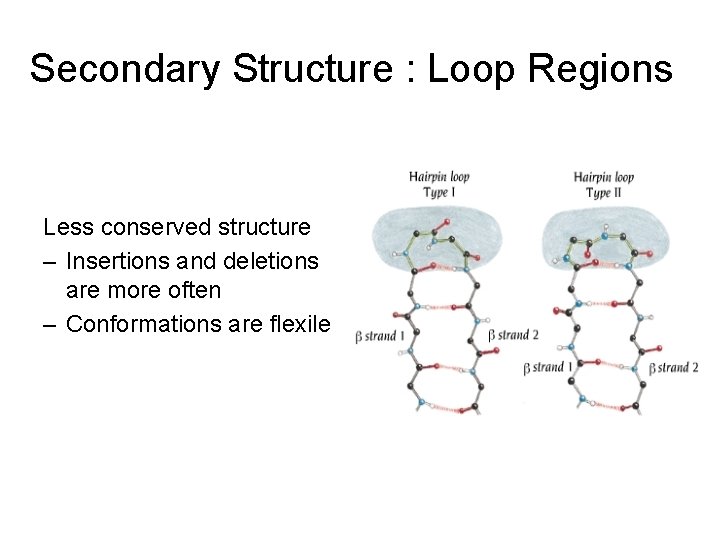
Secondary Structure : Loop Regions Less conserved structure – Insertions and deletions are more often – Conformations are flexile

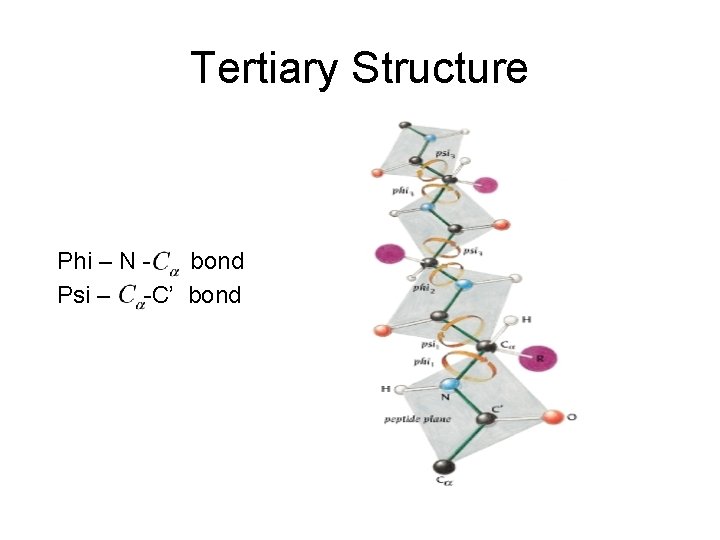
Tertiary Structure Phi – N bond Psi – -C’ bond

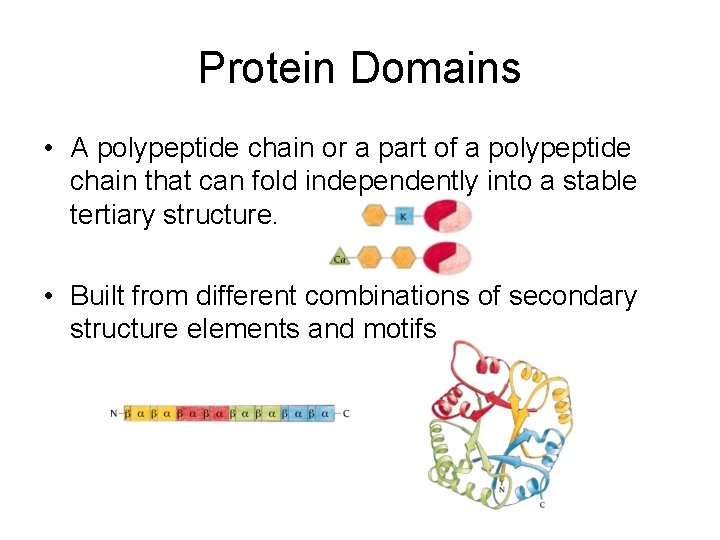
Protein Domains • A polypeptide chain or a part of a polypeptide chain that can fold independently into a stable tertiary structure. • Built from different combinations of secondary structure elements and motifs

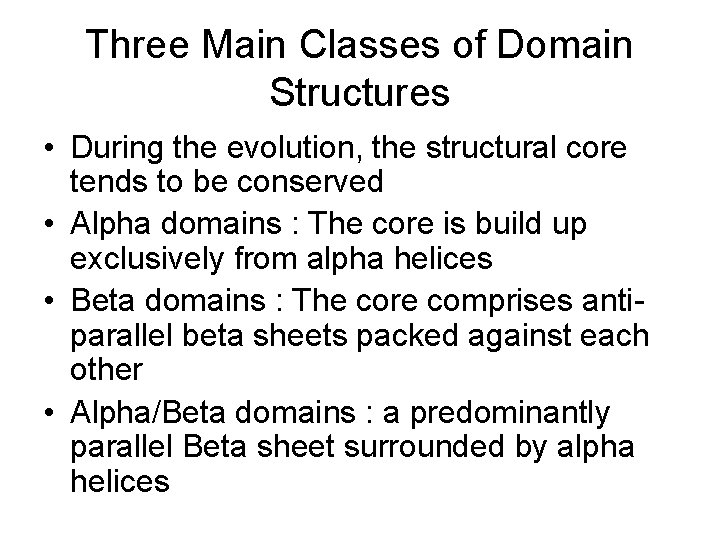
Three Main Classes of Domain Structures • During the evolution, the structural core tends to be conserved • Alpha domains : The core is build up exclusively from alpha helices • Beta domains : The core comprises antiparallel beta sheets packed against each other • Alpha/Beta domains : a predominantly parallel Beta sheet surrounded by alpha helices

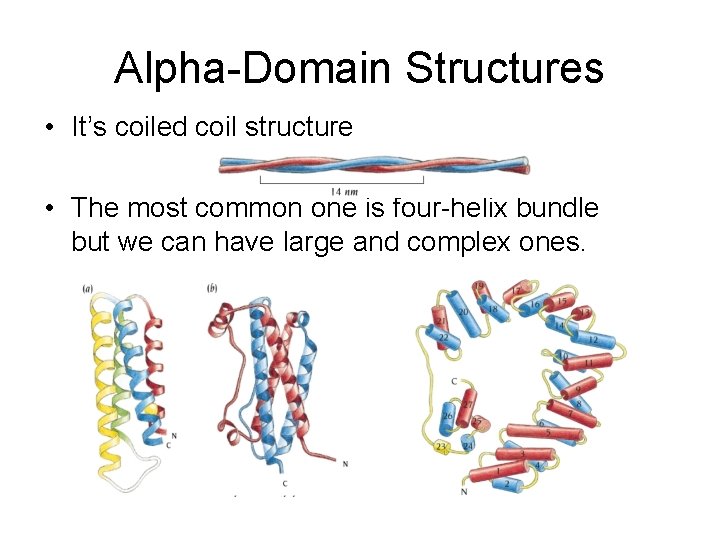
Alpha-Domain Structures • It’s coiled coil structure • The most common one is four-helix bundle but we can have large and complex ones.

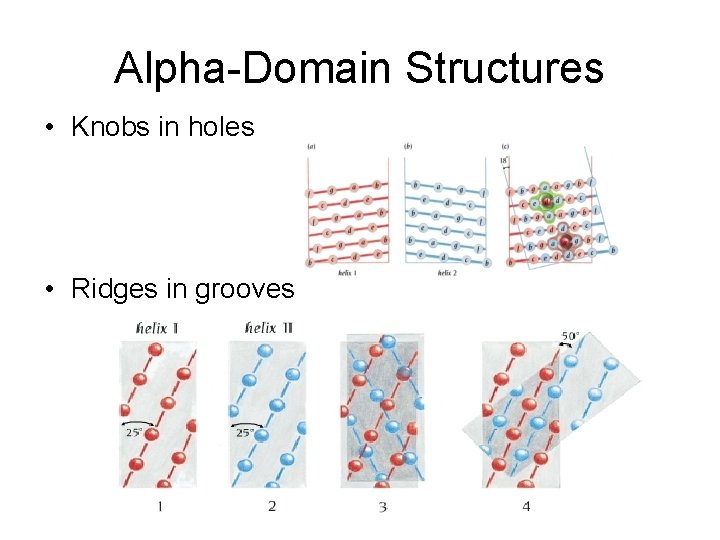
Alpha-Domain Structures • Knobs in holes • Ridges in grooves

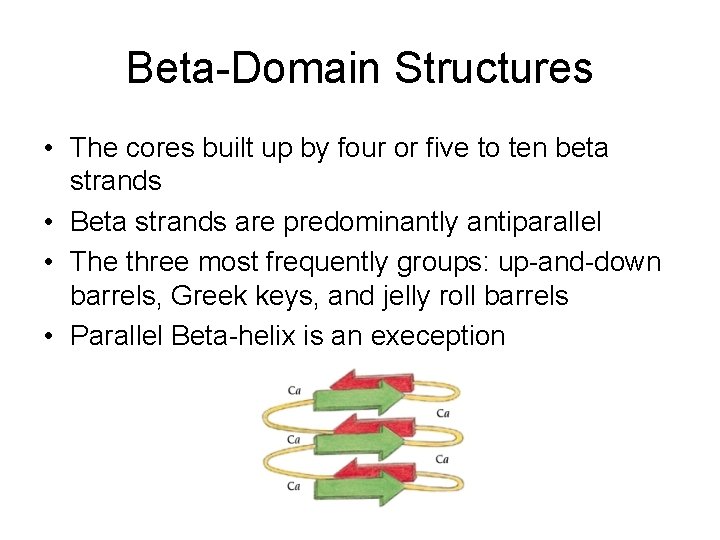
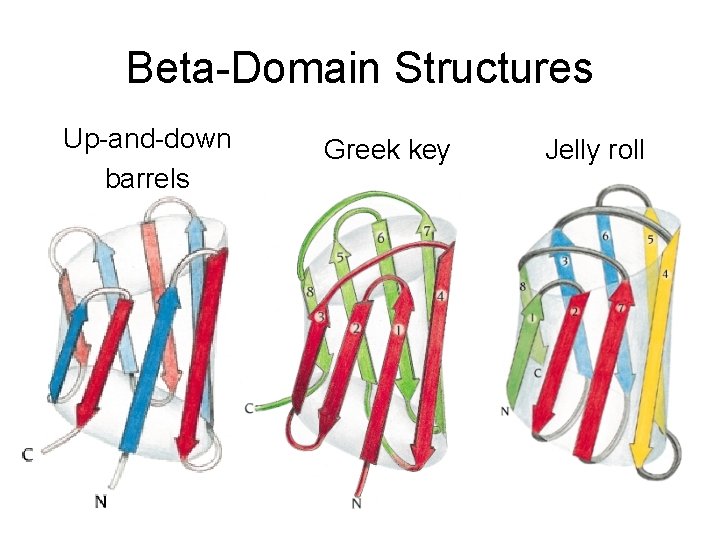
Beta-Domain Structures • The cores built up by four or five to ten beta strands • Beta strands are predominantly antiparallel • The three most frequently groups: up-and-down barrels, Greek keys, and jelly roll barrels • Parallel Beta-helix is an exeception

Beta-Domain Structures Up-and-down barrels Greek key Jelly roll

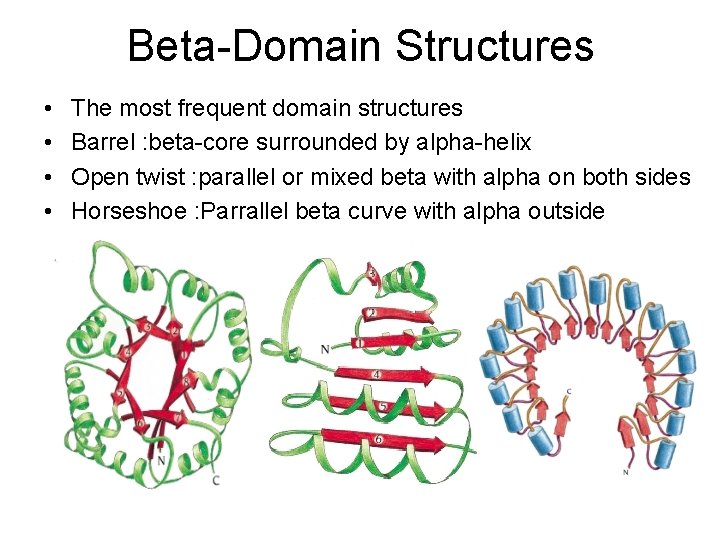
Beta-Domain Structures • • The most frequent domain structures Barrel : beta-core surrounded by alpha-helix Open twist : parallel or mixed beta with alpha on both sides Horseshoe : Parrallel beta curve with alpha outside

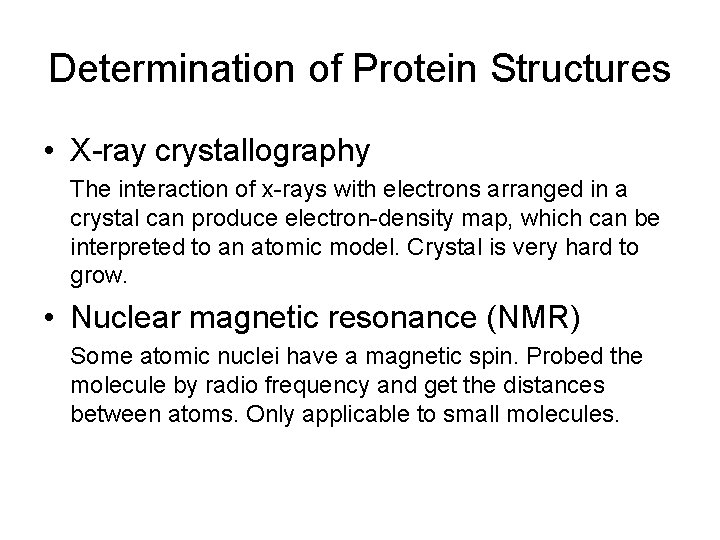
Determination of Protein Structures • X-ray crystallography The interaction of x-rays with electrons arranged in a crystal can produce electron-density map, which can be interpreted to an atomic model. Crystal is very hard to grow. • Nuclear magnetic resonance (NMR) Some atomic nuclei have a magnetic spin. Probed the molecule by radio frequency and get the distances between atoms. Only applicable to small molecules.
 Rui kuang
Rui kuang Namenode hdfs
Namenode hdfs Zhiming kuang
Zhiming kuang Hey hey bye bye
Hey hey bye bye Carrier vs channel proteins
Carrier vs channel proteins Protein-protein docking
Protein-protein docking Dont ask why why why
Dont ask why why why Poema de rui barbosa
Poema de rui barbosa Rui barbosa poemas
Rui barbosa poemas Poskrom dla koni wymiary
Poskrom dla koni wymiary Arianna rui
Arianna rui Rui seabra ferreira junior
Rui seabra ferreira junior Quản trị rủi ro và khủng hoảng
Quản trị rủi ro và khủng hoảng Rui dai wrds
Rui dai wrds Rui zhang unimelb
Rui zhang unimelb Rui
Rui Rui carlos botter
Rui carlos botter Rui carlos botter
Rui carlos botter