Genes and human history Gil Mc Vean Department





















- Slides: 43

Genes and human history Gil Mc. Vean, Department of Statistics, Oxford Contact: mcvean@stats. ox. ac. uk

2

3 • Where does the variation come from? • How old are the genetic differences between us? • Are these differences important?

How different are our genomes?

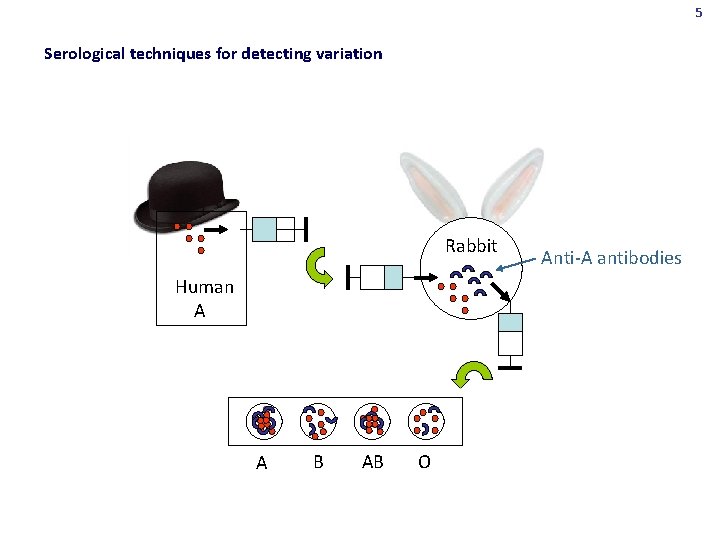
5 Serological techniques for detecting variation Rabbit Human A A B AB O Anti-A antibodies

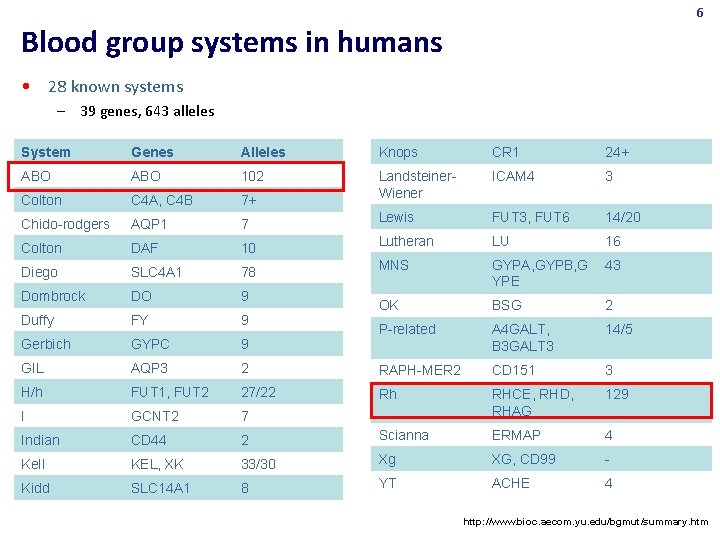
6 Blood group systems in humans • 28 known systems – 39 genes, 643 alleles System Genes Alleles Knops CR 1 24+ ABO 102 ICAM 4 3 Colton C 4 A, C 4 B 7+ Landsteiner. Wiener Chido-rodgers AQP 1 7 Lewis FUT 3, FUT 6 14/20 Colton DAF 10 Lutheran LU 16 Diego SLC 4 A 1 78 MNS 43 Dombrock DO 9 GYPA, GYPB, G YPE Duffy FY 9 OK BSG 2 Gerbich GYPC 9 P-related A 4 GALT, B 3 GALT 3 14/5 GIL AQP 3 2 RAPH-MER 2 CD 151 3 H/h FUT 1, FUT 2 27/22 Rh 129 I GCNT 2 7 RHCE, RHD, RHAG Indian CD 44 2 Scianna ERMAP 4 Kell KEL, XK 33/30 Xg XG, CD 99 - Kidd SLC 14 A 1 8 YT ACHE 4 http: //www. bioc. aecom. yu. edu/bgmut/summary. htm

7 Protein electroporesis • Changes in mass/charge ratio resulting from amino acid substitutions in proteins can be detected Starch or agar gel -- +-+ +- - + Direction of travel • In humans, about 30% of all loci show polymorphism with a 6% chance of a pair of randomly drawn alleles at a locus being different Lewontin and Hubby (1966) Harris(1966)

8 The rise of DNA sequencing GATAAGACGGTGATACTCACGCGACGGGCTTGGGCGCCGACTCGTTCAGACGGTGACCCAACTTATCCGATCGACCC CGGGTCCCGATTTAGACTCGGTATCATTTCTGGTGATTATCGCCTGCAGGTTCAAGAACACGTTTGCAGCAAGAAGT GAGGGATTTTGTCAGTGATCCCAGTCTACGGAGCCAGTCACCTCTGGTAGTGAAATTTTATTCGTTCATCTTCATAT AAGTCGCAGACCGCACGATGGGGGACAGAATACTCGCACAGGAAGAACCGCGATGAACCGAGGTAACCTAACATCCT AAGCCATTCCAACGAGGCTTTCGTAACCAAATCAGTTCTTCCCAGTCCAGATGAGGCGAACGTAGGTGCTGTTGGAA CCATGAGTGGCCAACAGAATACTGTGGATGCTAATGGAATGTGTTAATCAGACGTTTGCTGATGTGACACAT TGGTCGCTGCTCTTTGATGCGGAAATCTATGAGCGGTCAAACCGATACAAACCCGGCTATGTCGTTCGCACAACAGT CGGGTCCCACCCCATTGTTCTTATGAAGGTATTACTGGTCATACGATGCTTTTGCGACGCATCCCTATGACGA GAGTGCAGTCAGACCCCTCGACCATTTCCCTTAGAAAGACCACCCATCTCTTCAAAGTTATTCTCCGTGACATGCGA ACGCTGAAGGATAAGGAGCGGCATGCAGACTTTTATGTGTGCTCTCTGCTGGTCCAGCGGCATCTAAACGTCTCATC ACTAGGGCCACGCAGTCGTTTTTAAGAGGCTCTATTTTTACTAATTATTCTTGTCCACCACGACCTCTCAGCGCGGC AGATAGGTTCACAGGCTAGCGTCGGGTAATGCATTGCAGTTTCGTTACTCGTTCAGACAAGACTCGATGCTTTACAC TCACGACCCGCAAAGCCTTGGCCTTACAAGGGTATTAGGCCGAACACTTATCGCCGAAGGTACGTCGGCTATT GTAGCCCAAACCCTAGACTGAGCCCTAACCTCTACGCGTATCTTATAGGTTCAGAACGCCGAAGGACTATTCTCACG GCATTCATGGTTAAAAGAGAGTCGAGGCGCCTGCTATATGTGCCGAGTCCCATTAGTCAGTACACTTGCCATCACAT TTGTCCTGTTAGGCGGACACTTAGAGTAAGCGTACAACGCCTTACAACGAGACGCAGATCGCTTTTCTAATTGCGCC GCGTCTCTACCATCGTGGCCAGTTCATACTCACACGGAGGTGTGCAACCCGTAACACGAGTGCTCACTTTATA ATAAGTCAGCGTTCAGGACTGAGTGCAACCAATCTACGCCAGGAATCGCAAACAGCGCTCATAAACTTCTTACCTTT CCATAGCGCGCCTTTCGAGTATTATTGACCGTTAGGACTACGATAGGCTTCGACAATAGACCCTATCTGCGCATCAT TACCTCTCACCGGGGGAAATTCCAATCTGTCCAGGGCGCCCGTTTTTTTAAGACCTTAGTGCCCATGAACTGGCTCAAGCAATAGCGGCTGCTCGTGCCATGCGTGAGCTGGCGGCCAAATCGGACTCACGGACAAGTCTGC CCCCTTGTGAGTTAGTGTTGGCTTGACAACTCTAAAGTCCGAACCCATCGTGCGGCCATCCTACGTGGTGTAGCTTT GGCCCATAACCTGGTTACTCACTATCCTGCGACTCGTCTGGTCTCACTAGGCGATTCCCCCCGGCTTCGTATT GCAACATTCTAACGAATGCGAAGTCAAACAGTCCAGCTTAACAAAGGGGTCTTGACGAGACTCTGTAATCGTCTGCT AGCCCCGGACTCTGTTGTCGAAGGCAATTTGACGACCCACACGAGGTGCAGACGTAGTCAGGCCTGATAGCTATGTA TGCAGGCATATCCCTATAAAGTAGCGTTTGGTTATCCTACCATTAGCCGTTTCCGCATCTACCAGTGTCGACCGG

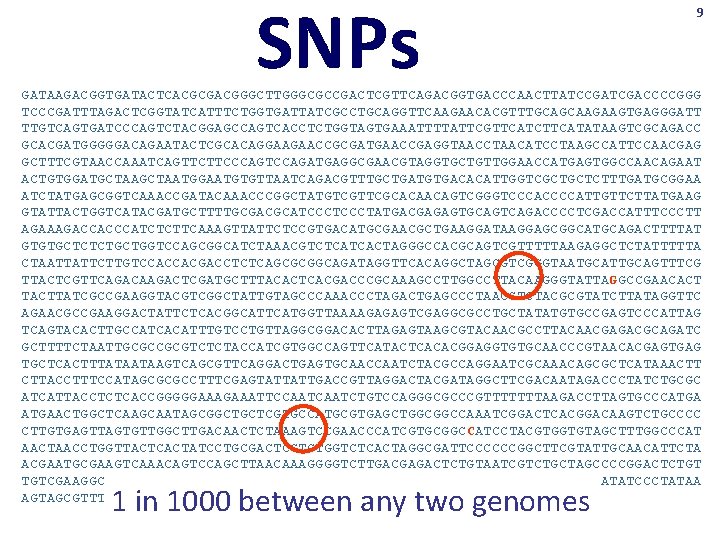
SNPs 9 GATAAGACGGTGATACTCACGCGACGGGCTTGGGCGCCGACTCGTTCAGACGGTGACCCAACTTATCCGATCGACCCCGGG TCCCGATTTAGACTCGGTATCATTTCTGGTGATTATCGCCTGCAGGTTCAAGAACACGTTTGCAGCAAGAAGTGAGGGATT TTGTCAGTGATCCCAGTCTACGGAGCCAGTCACCTCTGGTAGTGAAATTTTATTCGTTCATCTTCATATAAGTCGCAGACC GCACGATGGGGGACAGAATACTCGCACAGGAAGAACCGCGATGAACCGAGGTAACCTAACATCCTAAGCCATTCCAACGAG GCTTTCGTAACCAAATCAGTTCTTCCCAGTCCAGATGAGGCGAACGTAGGTGCTGTTGGAACCATGAGTGGCCAACAGAAT ACTGTGGATGCTAATGGAATGTGTTAATCAGACGTTTGCTGATGTGACACATTGGTCGCTGCTCTTTGATGCGGAA ATCTATGAGCGGTCAAACCGATACAAACCCGGCTATGTCGTTCGCACAACAGTCGGGTCCCACCCCATTGTTCTTATGAAG GTATTACTGGTCATACGATGCTTTTGCGACGCATCCCTATGACGAGAGTGCAGTCAGACCCCTCGACCATTTCCCTT AGAAAGACCACCCATCTCTTCAAAGTTATTCTCCGTGACATGCGAACGCTGAAGGATAAGGAGCGGCATGCAGACTTTTAT GTGTGCTCTCTGCTGGTCCAGCGGCATCTAAACGTCTCATCACTAGGGCCACGCAGTCGTTTTTAAGAGGCTCTATTTTTA CTAATTATTCTTGTCCACCACGACCTCTCAGCGCGGCAGATAGGTTCACAGGCTAGCGTCGGGTAATGCATTGCAGTTTCG TTACTCGTTCAGACAAGACTCGATGCTTTACACTCACGACCCGCAAAGCCTTGGCCTTACAAGGGTATTAGGCCGAACACT TACTTATCGCCGAAGGTACGTCGGCTATTGTAGCCCAAACCCTAGACTGAGCCCTAACCTCTACGCGTATCTTATAGGTTC AGAACGCCGAAGGACTATTCTCACGGCATTCATGGTTAAAAGAGAGTCGAGGCGCCTGCTATATGTGCCGAGTCCCATTAG TCAGTACACTTGCCATCACATTTGTCCTGTTAGGCGGACACTTAGAGTAAGCGTACAACGCCTTACAACGAGACGCAGATC GCTTTTCTAATTGCGCCGCGTCTCTACCATCGTGGCCAGTTCATACTCACACGGAGGTGTGCAACCCGTAACACGAGTGAG TGCTCACTTTATAATAAGTCAGCGTTCAGGACTGAGTGCAACCAATCTACGCCAGGAATCGCAAACAGCGCTCATAAACTT CTTACCTTTCCATAGCGCGCCTTTCGAGTATTATTGACCGTTAGGACTACGATAGGCTTCGACAATAGACCCTATCTGCGC ATCATTACCTCTCACCGGGGGAAATTCCAATCTGTCCAGGGCGCCCGTTTTTTTAAGACCTTAGTGCCCATGAACTGGCTCAAGCAATAGCGGCTGCTCGTGCCATGCGTGAGCTGGCGGCCAAATCGGACTCACGGACAAGTCTGCCCC CTTGTGAGTTAGTGTTGGCTTGACAACTCTAAAGTCCGAACCCATCGTGCGGCCATCCTACGTGGTGTAGCTTTGGCCCAT AACTAACCTGGTTACTCACTATCCTGCGACTCGTCTGGTCTCACTAGGCGATTCCCCCCGGCTTCGTATTGCAACATTCTA ACGAATGCGAAGTCAAACAGTCCAGCTTAACAAAGGGGTCTTGACGAGACTCTGTAATCGTCTGCTAGCCCCGGACTCTGT TGTCGAAGGCAATTTGACGACCCACACGAGGTGCAGACGTAGTCAGGCCTGATAGCTATGCAGGCATATCCCTATAA AGTAGCGTTTGGTTATCCTACCATTAGCCGTTTCCGCATCTACCAGTGTCGACCGG Single Nucleotide Polymorphisms TGCATTGCGTAGGC TGCATTCCGTAGGC 1 in 1000 between any two genomes

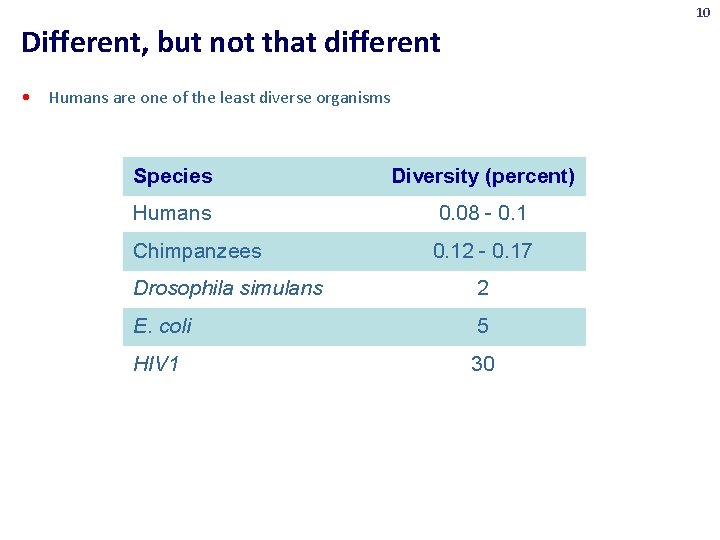
10 Different, but not that different • Humans are one of the least diverse organisms Species Diversity (percent) Humans 0. 08 - 0. 1 Chimpanzees 0. 12 - 0. 17 Drosophila simulans 2 E. coli 5 HIV 1 30

11 c. 3, 000 SNPs in 270 people

12 c. 40, 000 SNPs in 1000 people

13 How do we differ? – Let me count the ways • Single nucleotide polymorphisms TGCATTGCGTAGGC TGCATTCCGTAGGC • Short indels (=insertion/deletion) TGCATT---TAGGC TGCATTCCGTAGGC • Microsatellite (STR) repeat number TGCTCATCAGC TGCTCATCA------GC • Minisatellites ≤ 100 bp • Repeated genes – r. RNA, histones • Large inversions, deletions – Y chromosome, Copy Number Variants (CNVs) 1 -5 kb

14 Y chromosome variation • Non-pathological rearrangements of the AZFc region on the Y chromosome

15 Copy-number variation in genes • Variation in gene number can contribute to phenotypic variation Perry et al. 2007

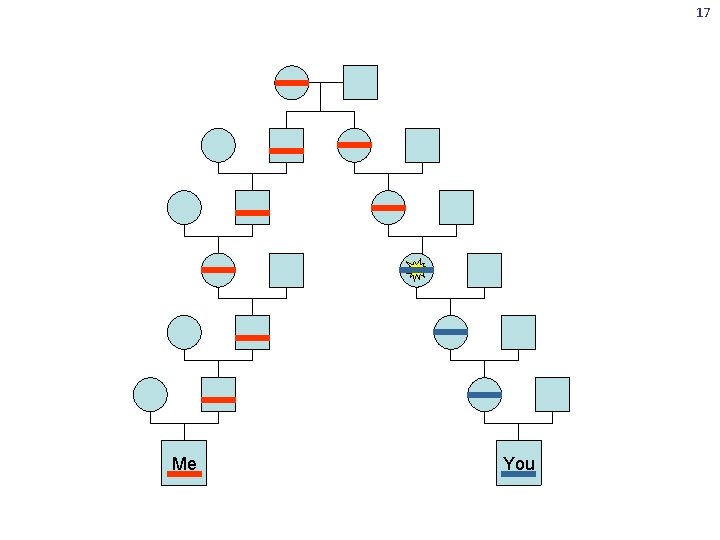
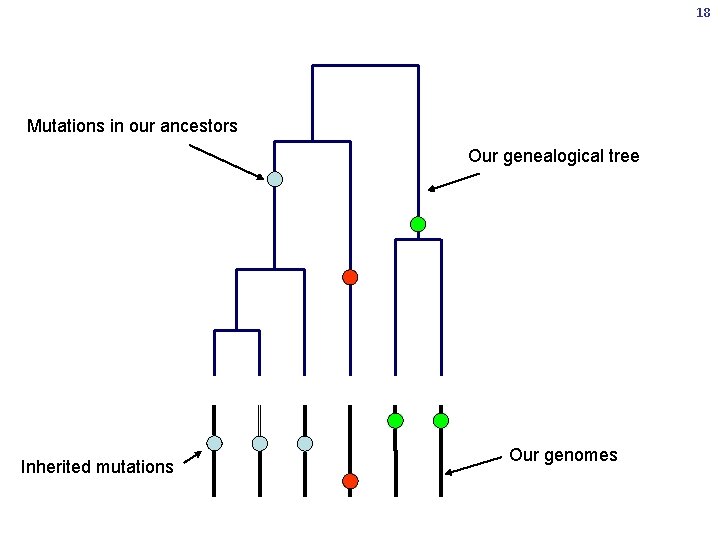
16 Where does genetic variation come from? • You will pass on about 60 new mutations to each of your children • Most of these are destined to die out within a few generations • Most variation is inherited from our ancestors

17 Me You

18 Mutations in our ancestors Our genealogical tree Inherited mutations Our genomes

19 mt. DNA Eve Vigilant et al. (1991)

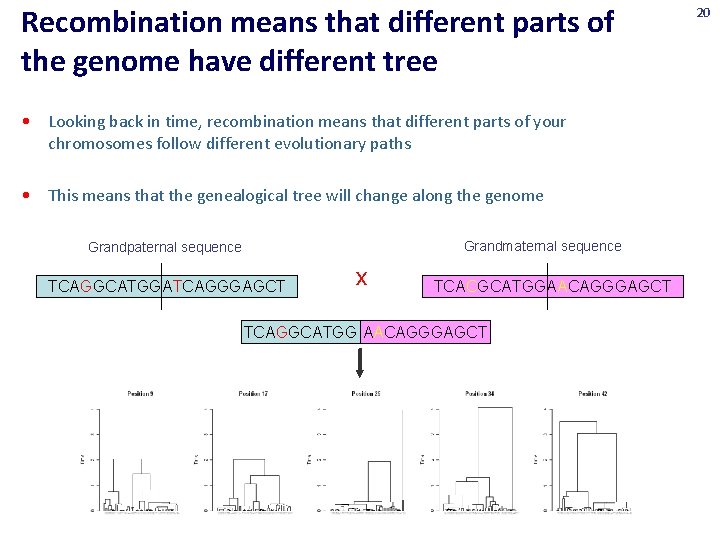
Recombination means that different parts of the genome have different tree • Looking back in time, recombination means that different parts of your chromosomes follow different evolutionary paths • This means that the genealogical tree will change along the genome Grandmaternal sequence Grandpaternal sequence TCAGGCATGGATCAGGGAGCT x TCACGCATGGAACAGGGAGCT TCAGGCATGG AACAGGGAGCT 20

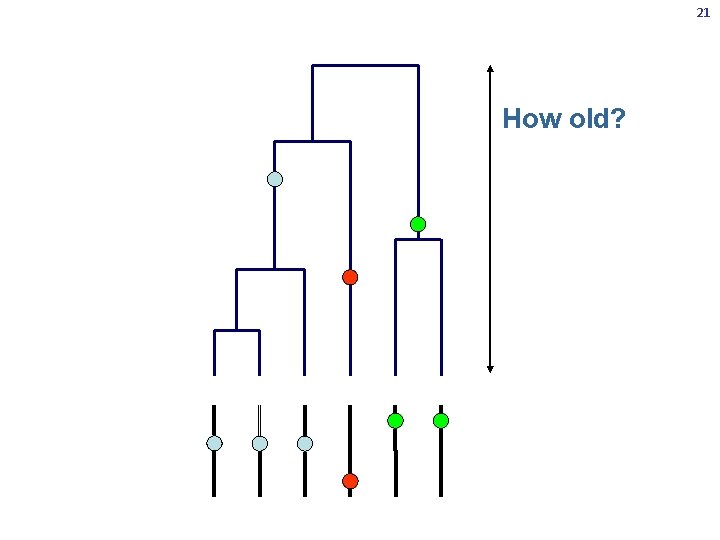
21 How old?

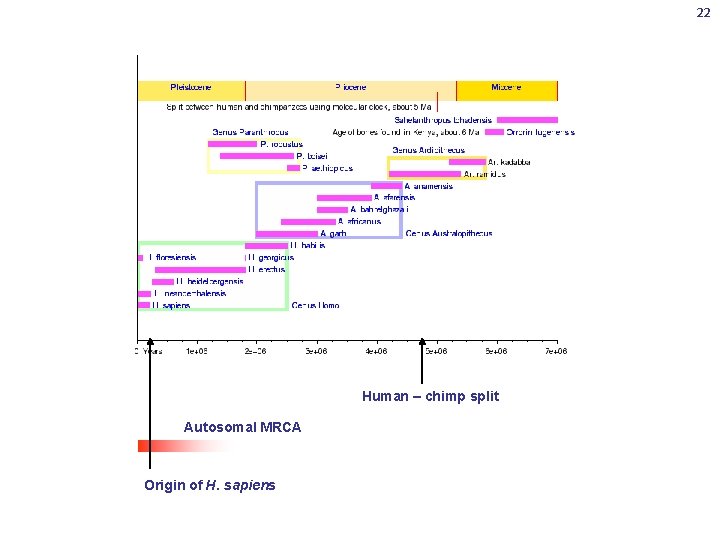
22 Human – chimp split Autosomal MRCA Origin of H. sapiens

Homo erectus

Australopithecus afarensis

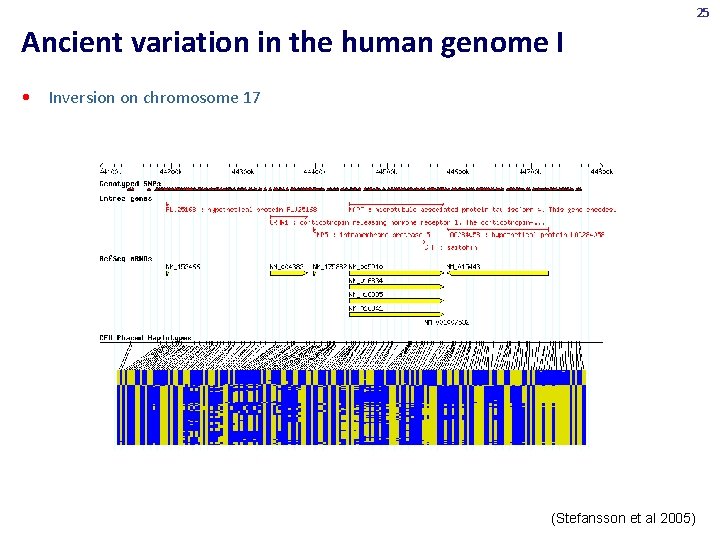
25 Ancient variation in the human genome I • Inversion on chromosome 17 (Stefansson et al 2005)

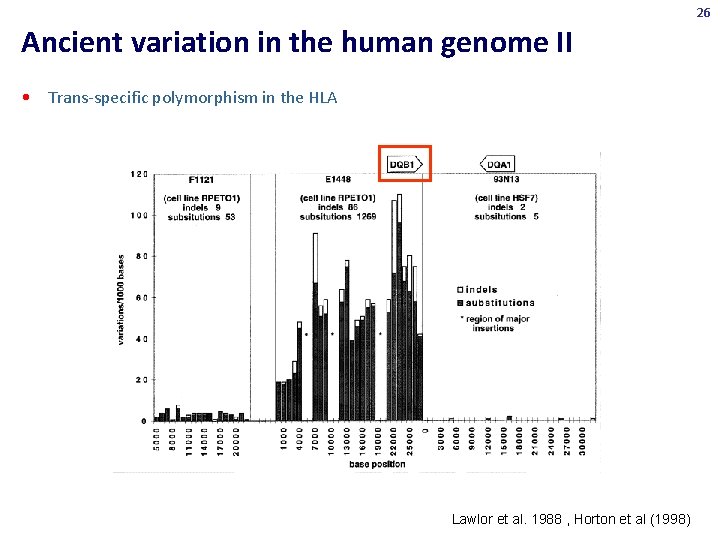
26 Ancient variation in the human genome II • Trans-specific polymorphism in the HLA Lawlor et al. 1988 , Horton et al (1998)

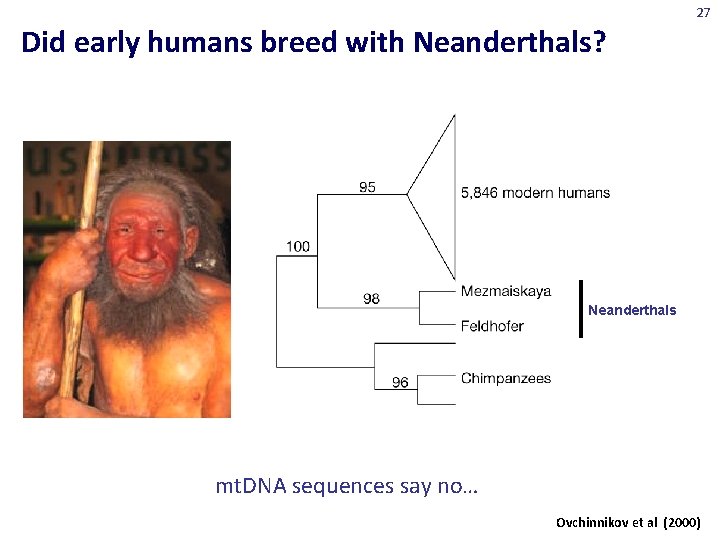
27 Did early humans breed with Neanderthals? Neanderthals mt. DNA sequences say no… Ovchinnikov et al (2000)

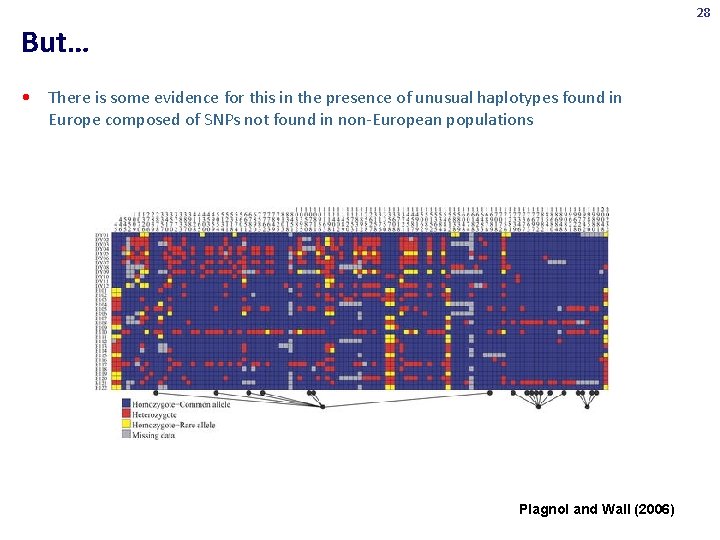
28 But… • There is some evidence for this in the presence of unusual haplotypes found in Europe composed of SNPs not found in non-European populations Plagnol and Wall (2006)

What are the genetic differences that make us human?

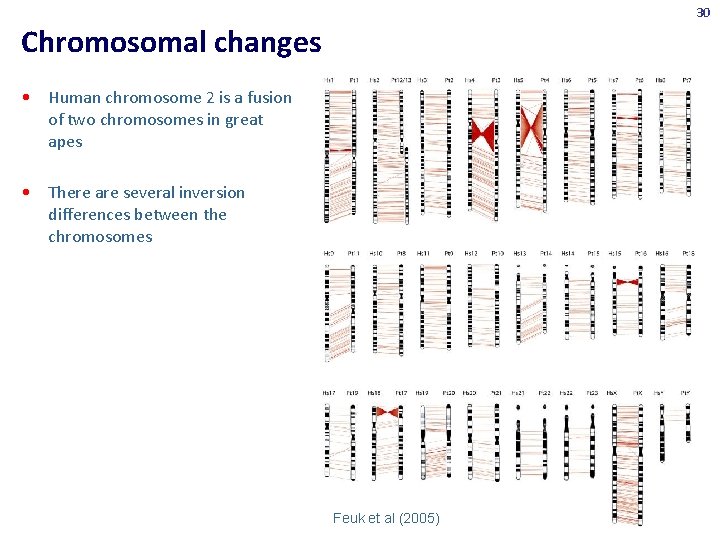
30 Chromosomal changes • Human chromosome 2 is a fusion of two chromosomes in great apes • There are several inversion differences between the chromosomes Feuk et al (2005)

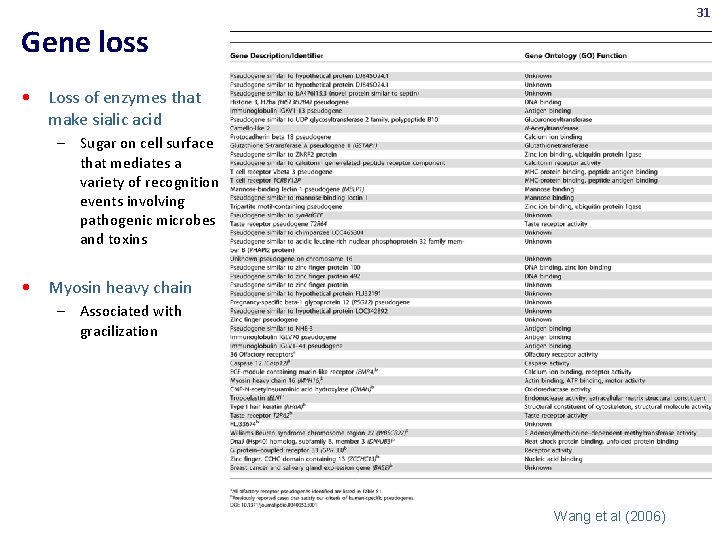
31 Gene loss • Loss of enzymes that make sialic acid – Sugar on cell surface that mediates a variety of recognition events involving pathogenic microbes and toxins • Myosin heavy chain – Associated with gracilization Wang et al (2006)

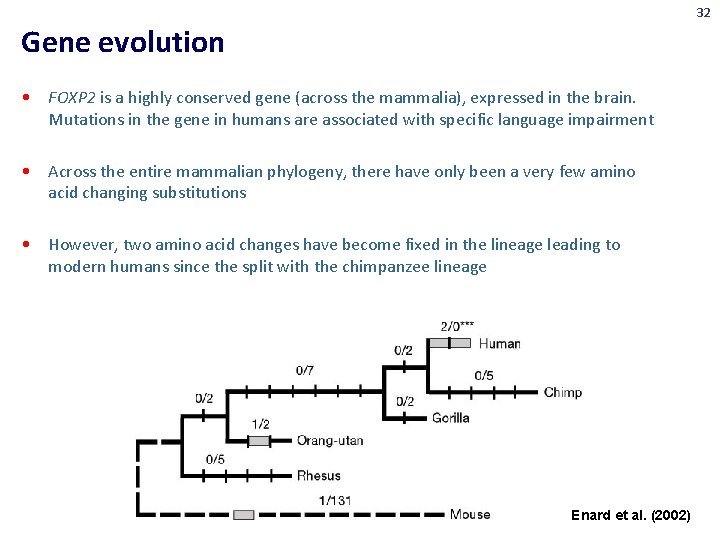
32 Gene evolution • FOXP 2 is a highly conserved gene (across the mammalia), expressed in the brain. Mutations in the gene in humans are associated with specific language impairment • Across the entire mammalian phylogeny, there have only been a very few amino acid changing substitutions • However, two amino acid changes have become fixed in the lineage leading to modern humans since the split with the chimpanzee lineage Enard et al. (2002)

Are the genetic differences between people and peoples important?

34 Infectious disease Diet Genome ? Physical environment Mating success

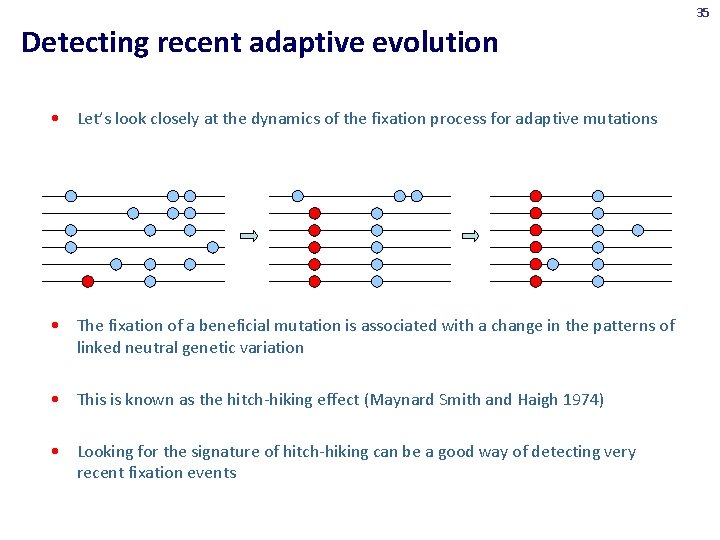
35 Detecting recent adaptive evolution • Let’s look closely at the dynamics of the fixation process for adaptive mutations • The fixation of a beneficial mutation is associated with a change in the patterns of linked neutral genetic variation • This is known as the hitch-hiking effect (Maynard Smith and Haigh 1974) • Looking for the signature of hitch-hiking can be a good way of detecting very recent fixation events

36 Lactose persistence

37 Lactose intolerance

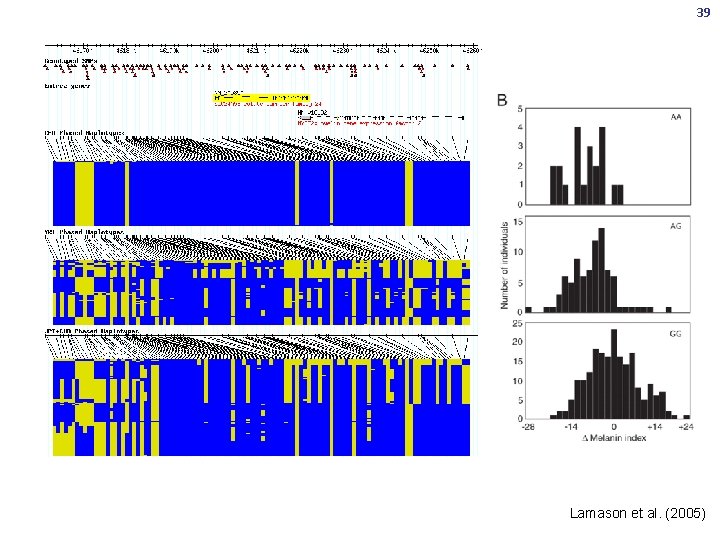
38 Skin pigmentation

39 Lamason et al. (2005)

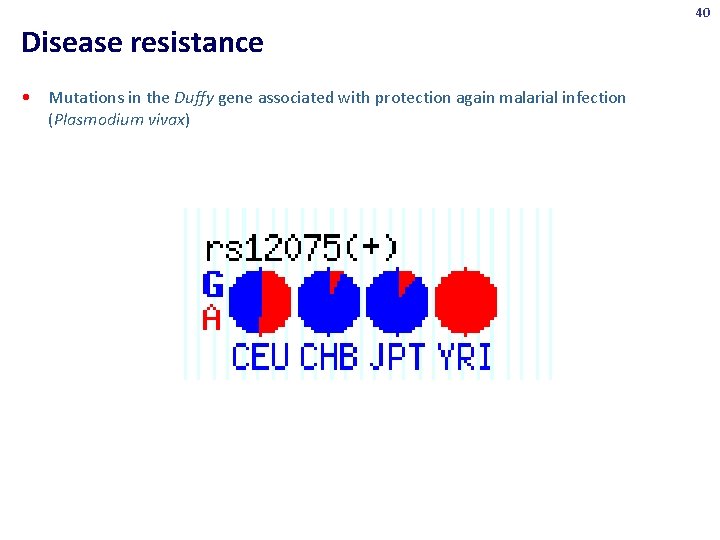
40 Disease resistance • Mutations in the Duffy gene associated with protection again malarial infection (Plasmodium vivax)

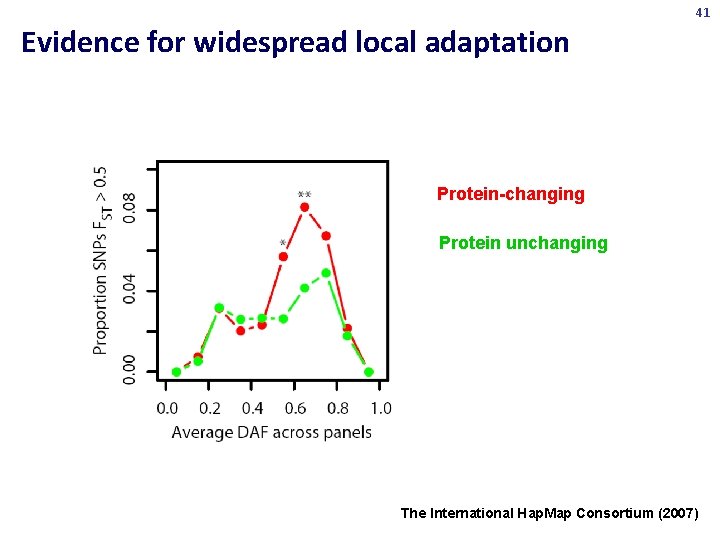
41 Evidence for widespread local adaptation Protein-changing Protein unchanging The International Hap. Map Consortium (2007)

42 Classes of selected genes Voight et al. (2005)

43 Reading • Human genetic variation – – – • The origin of modern humans – – – • Rosenberg et al. Genetic structure of human populations. Science 2002, 298: 2381 -2385. Conrad et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nature Genet. 2006, 1251 -1260. Mc. Vean et al. Perspectives on human genetic variation from the International Hap. Map Project. PLo. S Genetics 2005, 1: e 54. Reed & Tishkoff. African human diversity, origins and migrations. Curr Opin Genet Dev. 2006 16: 597 -605. Jobling et al. Human evolutionary genetics: origins, peoples, and disease. Garland Science, 2004. Harding & Mc. Vean. A structured ancestral population for the evolution of modern humans. Curr. Op. Genet. Dev. 2004, 14: 667 -674. Natural selection – – – Lamason et al. SLC 24 A 5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 2005, 310: 1782 -1786. Sabeti et al. Positive natural selection in the human lineage. Science 2006, 312: 1614 -1620. Tishkoff et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007 39: 31 -40
 Human vean
Human vean Linked genes and unlinked genes
Linked genes and unlinked genes Polygenic inheritance
Polygenic inheritance Tga vean
Tga vean Poemas a la madre naturaleza
Poemas a la madre naturaleza Homeotic genes vs hox genes
Homeotic genes vs hox genes Iowa department of health and human services
Iowa department of health and human services Milwaukee health and human services
Milwaukee health and human services Maine department of health and human services
Maine department of health and human services Gil kalai quantum computing
Gil kalai quantum computing Gil kalai quantum computing
Gil kalai quantum computing Haskell kirkpatrick
Haskell kirkpatrick Pesquisa por imagem
Pesquisa por imagem Michelli gil jaimez
Michelli gil jaimez Forensic science serial killers
Forensic science serial killers Redondilha maior e menor
Redondilha maior e menor Imagen de gil gonzalez davila
Imagen de gil gonzalez davila Gil cuadros
Gil cuadros Robledo lima gil
Robledo lima gil Gil kirkpatrick
Gil kirkpatrick Rizal volunteered as military doctor in cuba
Rizal volunteered as military doctor in cuba Heart structure
Heart structure Auto pastoril del nacimiento gil vicente
Auto pastoril del nacimiento gil vicente Nelson javier duenas gil
Nelson javier duenas gil Comico de caracter farsa de ines pereira
Comico de caracter farsa de ines pereira O velho da horta personagens
O velho da horta personagens Javier riancho zarrabeitia
Javier riancho zarrabeitia Jaime gil aluja
Jaime gil aluja Lorenzo gil macia
Lorenzo gil macia Jonathan gil porn
Jonathan gil porn Name of the student
Name of the student Gil naor
Gil naor Ibeps
Ibeps Espaço de estados
Espaço de estados Roberto gil annes da silva
Roberto gil annes da silva Vicente gil agresion
Vicente gil agresion Gil, antonio carlos. como elaborar projetos de pesquisa.
Gil, antonio carlos. como elaborar projetos de pesquisa. Antonio gil
Antonio gil Gil gallegos
Gil gallegos Que es maximato
Que es maximato Qcia goals
Qcia goals Vicente gil agresor
Vicente gil agresor Gil 1999
Gil 1999 Infancia y confesiones
Infancia y confesiones