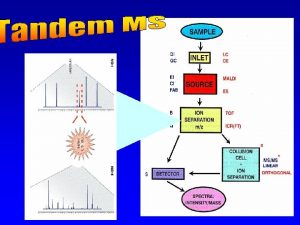
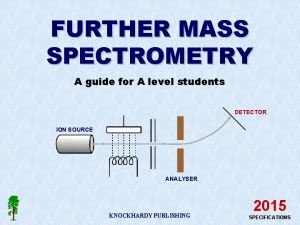
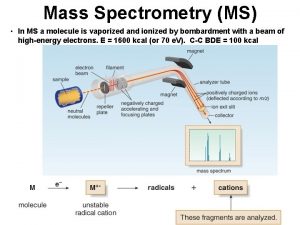
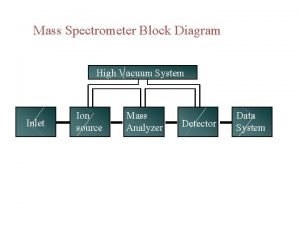
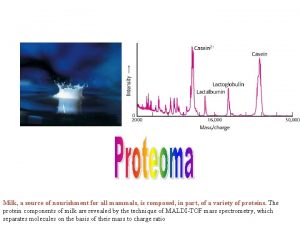
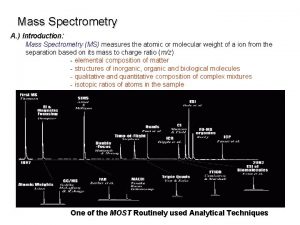
CSE 182 L 12 Mass Spectrometry Peptide identification



![D. P. forbidden pairs • Consider all pairs u, v – m[u] <= M/2, D. P. forbidden pairs • Consider all pairs u, v – m[u] <= M/2,](https://slidetodoc.com/presentation_image_h2/80626fd21d6485be320a03bad227c3ba/image-17.jpg)







- Slides: 43

CSE 182 -L 12 Mass Spectrometry Peptide identification Fa 06 CSE 182

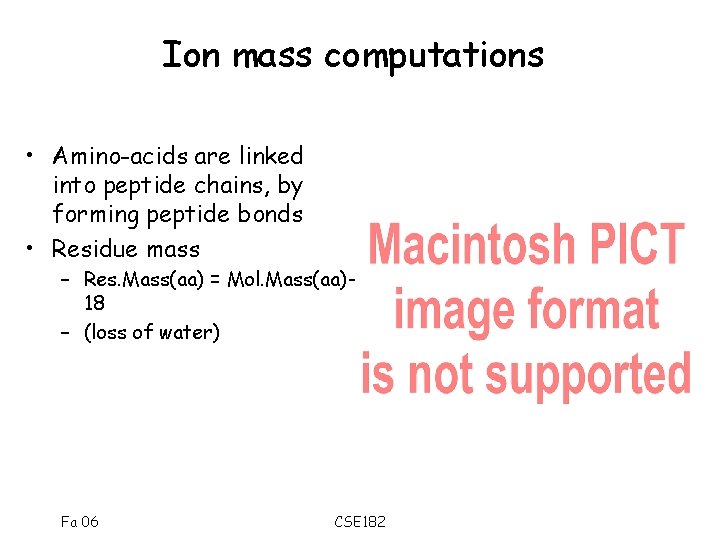
Ion mass computations • Amino-acids are linked into peptide chains, by forming peptide bonds • Residue mass – Res. Mass(aa) = Mol. Mass(aa)18 – (loss of water) Fa 06 CSE 182

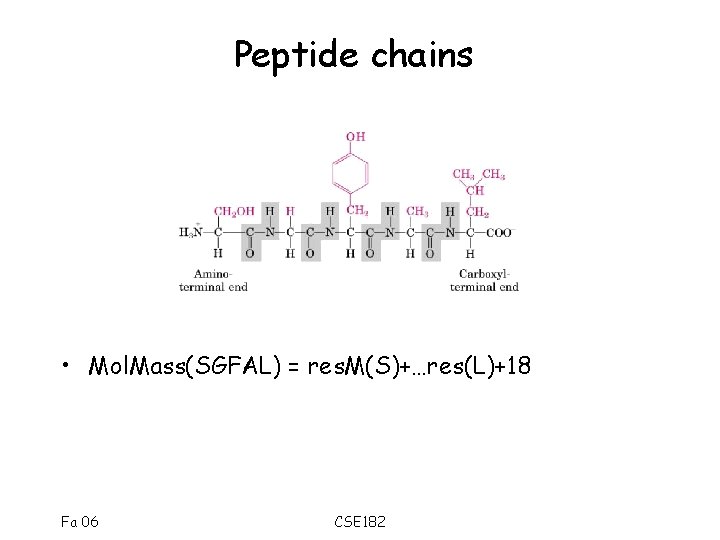
Peptide chains • Mol. Mass(SGFAL) = res. M(S)+…res(L)+18 Fa 06 CSE 182

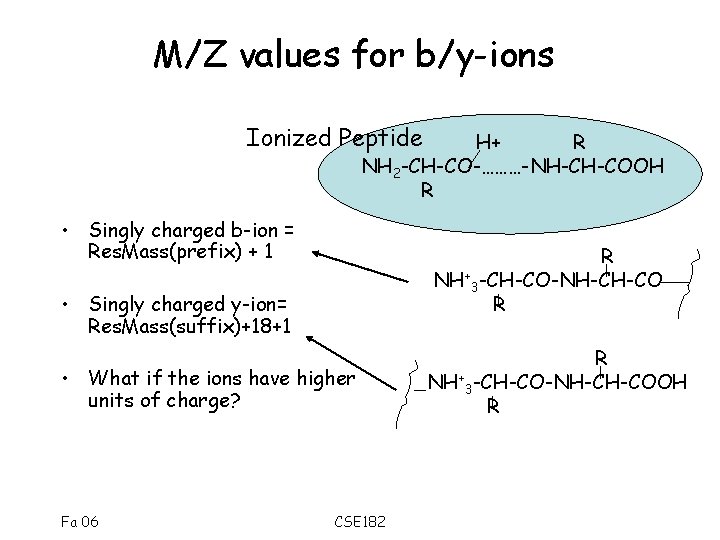
M/Z values for b/y-ions Ionized Peptide H+ R NH 2 -CH-CO-………-NH-CH-COOH R • Singly charged b-ion = Res. Mass(prefix) + 1 R NH+3 -CH-CO-NH-CH-CO R • Singly charged y-ion= Res. Mass(suffix)+18+1 • What if the ions have higher units of charge? Fa 06 CSE 182 R NH+3 -CH-CO-NH-CH-COOH R

De novo interpretation • Given a spectrum (a collection of b-y ions), compute the peptide that generated the spectrum. • A database of peptides is not given! • Useful? – Many genomes have not been sequenced, but are very useful. – Tagging/filtering – PTMs Fa 06 CSE 182

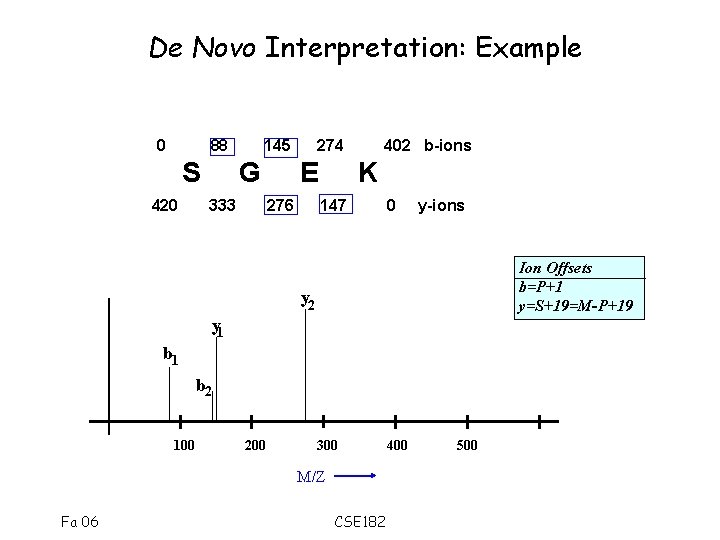
De Novo Interpretation: Example 0 88 S 420 145 G 333 274 E 276 402 b-ions K 147 0 y-ions Ion Offsets b=P+1 y=S+19=M-P+19 y 2 y 1 b 2 100 200 300 M/Z Fa 06 CSE 182 400 500

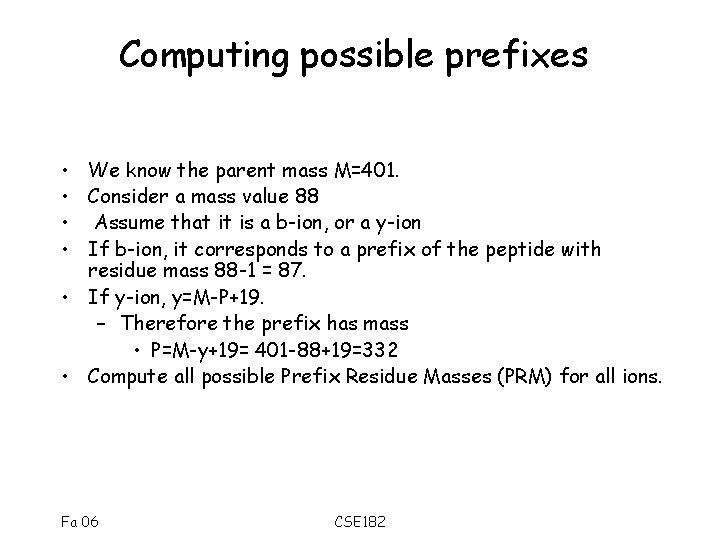
Computing possible prefixes • We know the parent mass M=401. • Consider a mass value 88 • Assume that it is a b-ion, or a y-ion • If b-ion, it corresponds to a prefix of the peptide with residue mass 88 -1 = 87. • If y-ion, y=M-P+19. – Therefore the prefix has mass • P=M-y+19= 401 -88+19=332 • Compute all possible Prefix Residue Masses (PRM) for all ions. Fa 06 CSE 182

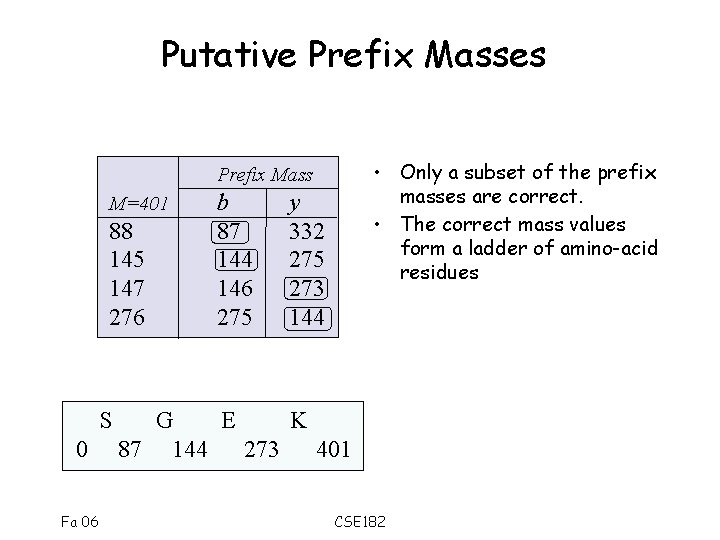
Putative Prefix Masses • Only a subset of the prefix masses are correct. • The correct mass values form a ladder of amino-acid residues Prefix Mass M=401 88 145 147 276 S 0 Fa 06 b 87 144 146 275 y 332 275 273 144 G E K 87 144 273 401 CSE 182

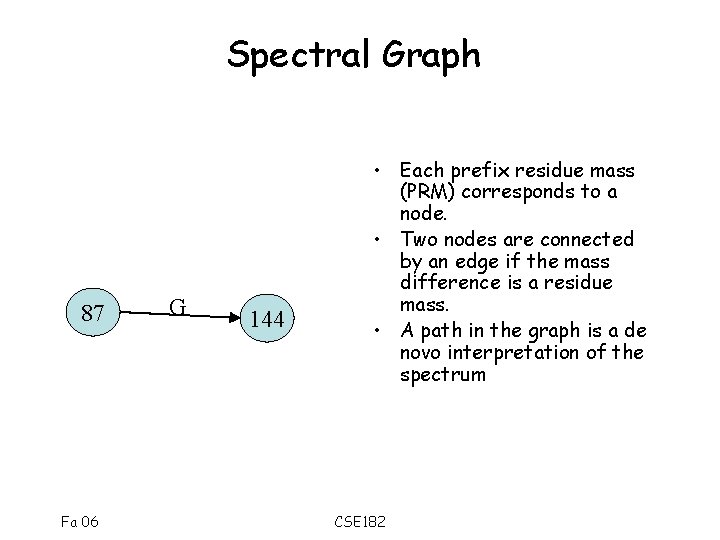
Spectral Graph 87 Fa 06 G 144 • Each prefix residue mass (PRM) corresponds to a node. • Two nodes are connected by an edge if the mass difference is a residue mass. • A path in the graph is a de novo interpretation of the spectrum CSE 182

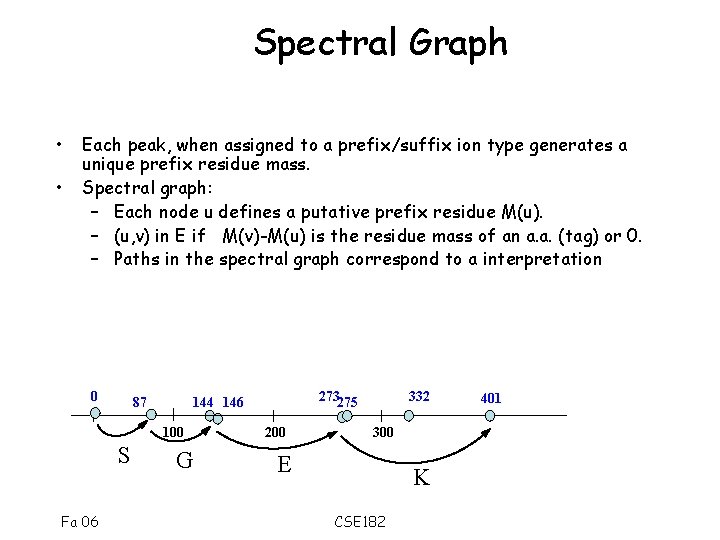
Spectral Graph • • Each peak, when assigned to a prefix/suffix ion type generates a unique prefix residue mass. Spectral graph: – Each node u defines a putative prefix residue M(u). – (u, v) in E if M(v)-M(u) is the residue mass of an a. a. (tag) or 0. – Paths in the spectral graph correspond to a interpretation 0 87 100 S Fa 06 273275 144 146 G 200 332 300 E K CSE 182 401

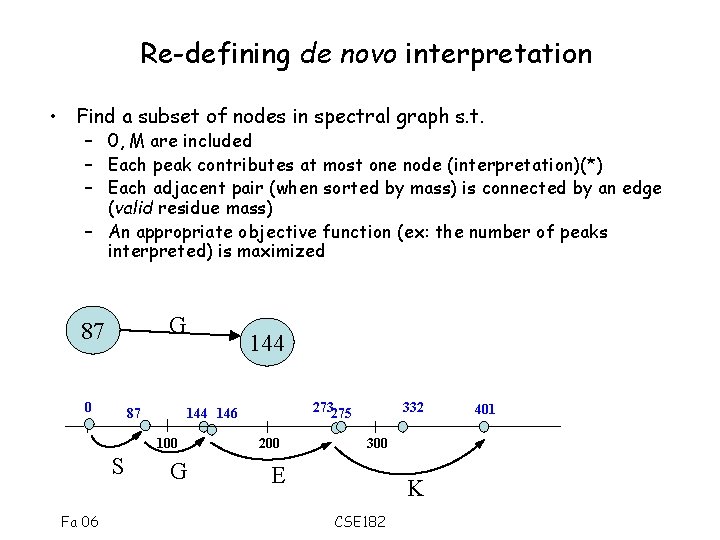
Re-defining de novo interpretation • Find a subset of nodes in spectral graph s. t. – 0, M are included – Each peak contributes at most one node (interpretation)(*) – Each adjacent pair (when sorted by mass) is connected by an edge (valid residue mass) – An appropriate objective function (ex: the number of peaks interpreted) is maximized G 87 0 87 Fa 06 273275 144 146 100 S 144 G 200 332 300 E K CSE 182 401

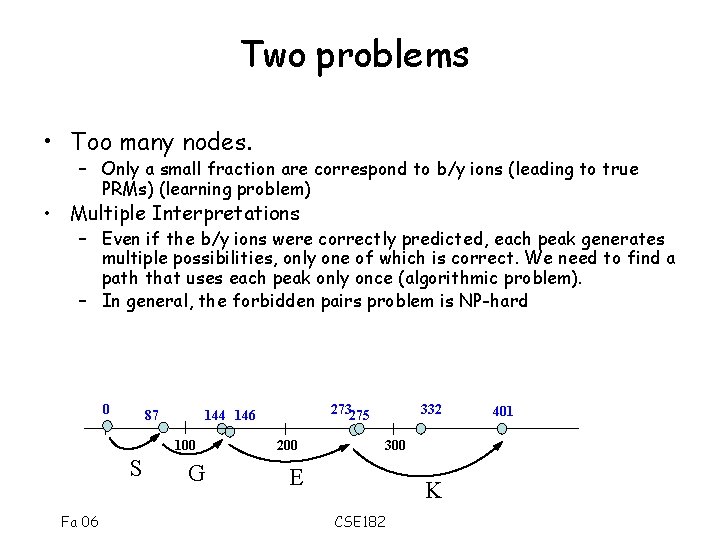
Two problems • Too many nodes. – Only a small fraction are correspond to b/y ions (leading to true PRMs) (learning problem) • Multiple Interpretations – Even if the b/y ions were correctly predicted, each peak generates multiple possibilities, only one of which is correct. We need to find a path that uses each peak only once (algorithmic problem). – In general, the forbidden pairs problem is NP-hard 0 87 100 S Fa 06 273275 144 146 G 200 332 300 E K CSE 182 401

Too many nodes • We will use other properties to decide if a peak is a b-y peak or not. • For now, assume that (u) is a score function for a peak u being a b-y ion. Fa 06 CSE 182

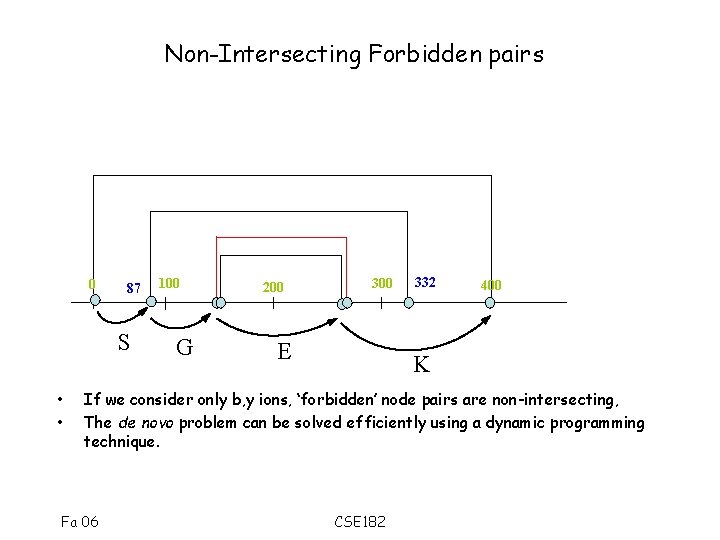
Multiple Interpretation • Each peak generates multiple possibilities, only one of which is correct. We need to find a path that uses each peak only once (algorithmic problem). • In general, the forbidden pairs problem is NP-hard • However, The b, y ions have a special noninterleaving property • Consider pairs (b 1, y 1), (b 2, y 2) – If (b 1 < b 2), then y 1 > y 2 Fa 06 CSE 182

Non-Intersecting Forbidden pairs 0 87 S • • 100 G 200 300 E 332 400 K If we consider only b, y ions, ‘forbidden’ node pairs are non-intersecting, The de novo problem can be solved efficiently using a dynamic programming technique. Fa 06 CSE 182

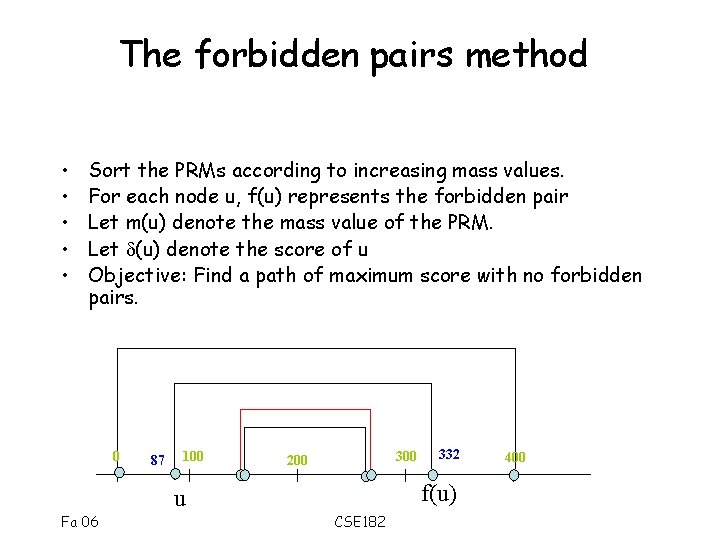
The forbidden pairs method • • • Sort the PRMs according to increasing mass values. For each node u, f(u) represents the forbidden pair Let m(u) denote the mass value of the PRM. Let (u) denote the score of u Objective: Find a path of maximum score with no forbidden pairs. 0 87 100 300 200 f(u) u Fa 06 332 CSE 182 400
![D P forbidden pairs Consider all pairs u v mu M2 D. P. forbidden pairs • Consider all pairs u, v – m[u] <= M/2,](https://slidetodoc.com/presentation_image_h2/80626fd21d6485be320a03bad227c3ba/image-17.jpg)
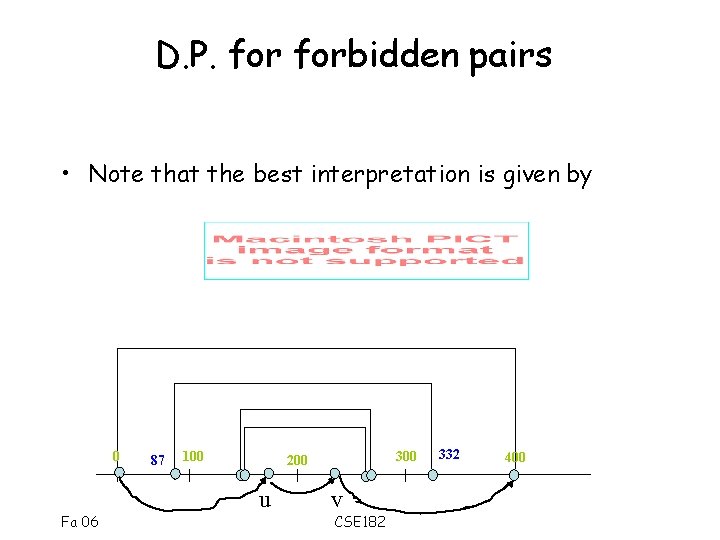
D. P. forbidden pairs • Consider all pairs u, v – m[u] <= M/2, m[v] >M/2 • Define S(u, v) as the best score of a forbidden pair path from – 0 ->u, and v->M • Is it sufficient to compute S(u, v) for all u, v? 0 87 100 300 200 u Fa 06 332 400 v CSE 182

D. P. forbidden pairs • Note that the best interpretation is given by 0 Fa 06 87 100 300 200 u v CSE 182 332 400

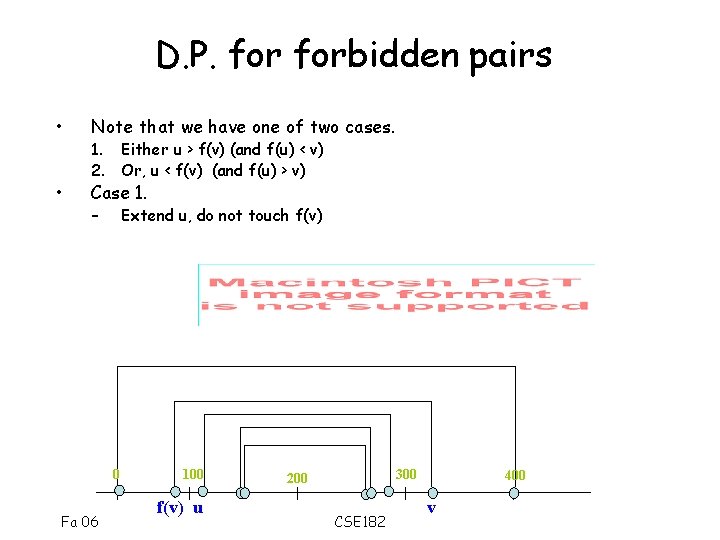
D. P. forbidden pairs • Note that we have one of two cases. • Case 1. Either u > f(v) (and f(u) < v) 2. Or, u < f(v) (and f(u) > v) – Extend u, do not touch f(v) 0 Fa 06 100 f(v) u 300 200 CSE 182 400 v

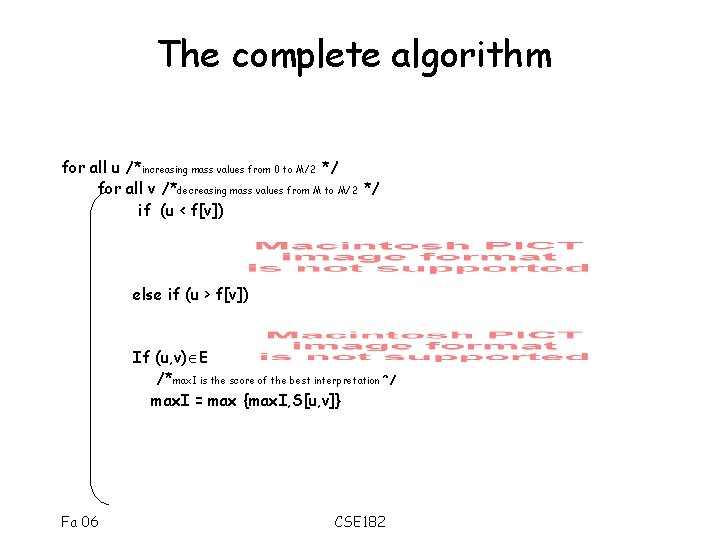
The complete algorithm for all u /*increasing mass values from 0 to M/2 */ for all v /*decreasing mass values from M to M/2 */ if (u < f[v]) else if (u > f[v]) If (u, v) E /*max. I is the score of the best interpretation*/ max. I = max {max. I, S[u, v]} Fa 06 CSE 182

De Novo: Second issue • Given only b, y ions, a forbidden pairs path will solve the problem. • However, recall that there are MANY other ion types. – – Fa 06 Typical length of peptide: 15 Typical # peaks? 50 -150? #b/y ions? Most ions are “Other” • a ions, neutral losses, isotopic peaks…. CSE 182

De novo: Weighting nodes in Spectrum Graph • Factors determining if the ion is b or y – Intensity (A large fraction of the most intense peaks are b or y) – Support ions – Isotopic peaks Fa 06 CSE 182

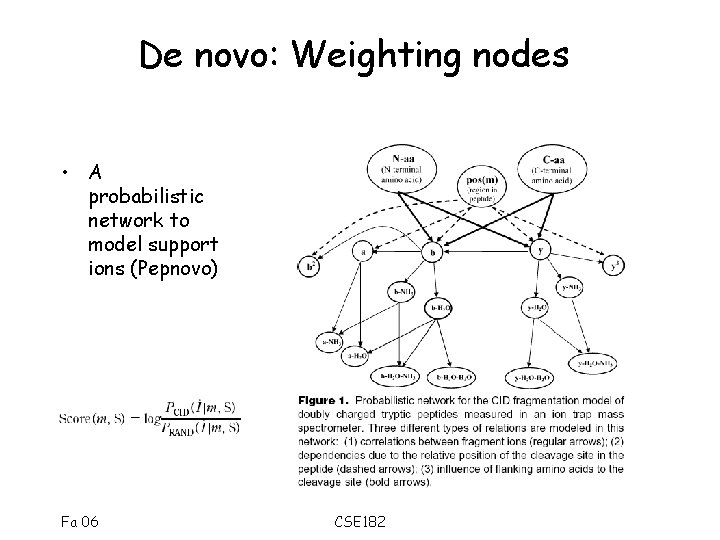
De novo: Weighting nodes • A probabilistic network to model support ions (Pepnovo) Fa 06 CSE 182

De Novo Interpretation Summary • The main challenge is to separate b/y ions from everything else (weighting nodes), and separating the prefix ions from the suffix ions (Forbidden Pairs). • As always, the abstract idea must be supplemented with many details. – – Noise peaks, incomplete fragmentation In reality, a PRM is first scored on its likelihood of being correct, and the forbidden pair method is applied subsequently. • In spite of these algorithms, de novo identification remains an error-prone process. When the peptide is in the database, db search is the method of choice. Fa 06 CSE 182

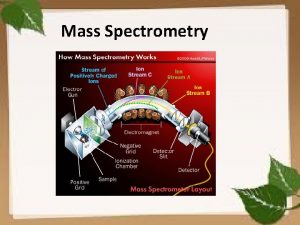
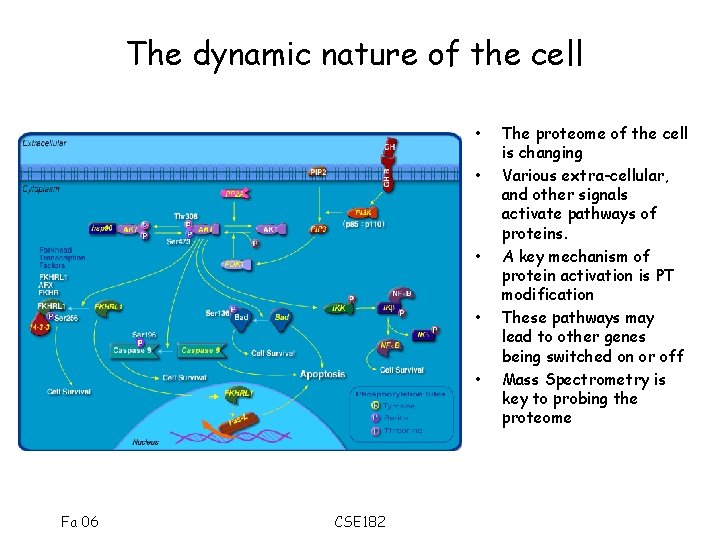
The dynamic nature of the cell • • • Fa 06 CSE 182 The proteome of the cell is changing Various extra-cellular, and other signals activate pathways of proteins. A key mechanism of protein activation is PT modification These pathways may lead to other genes being switched on or off Mass Spectrometry is key to probing the proteome

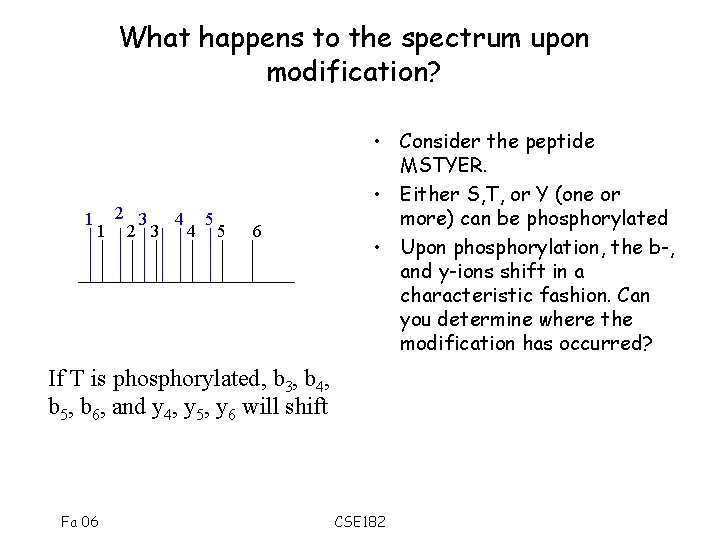
What happens to the spectrum upon modification? 1 2 3 4 5 6 • Consider the peptide MSTYER. • Either S, T, or Y (one or more) can be phosphorylated • Upon phosphorylation, the b-, and y-ions shift in a characteristic fashion. Can you determine where the modification has occurred? If T is phosphorylated, b 3, b 4, b 5, b 6, and y 4, y 5, y 6 will shift Fa 06 CSE 182

Effect of PT modifications on identification • The shifts do not affect de novo interpretation too much. Why? • Database matching algorithms are affected, and must be changed. • Given a candidate peptide, and a spectrum, can you identify the sites of modifications Fa 06 CSE 182

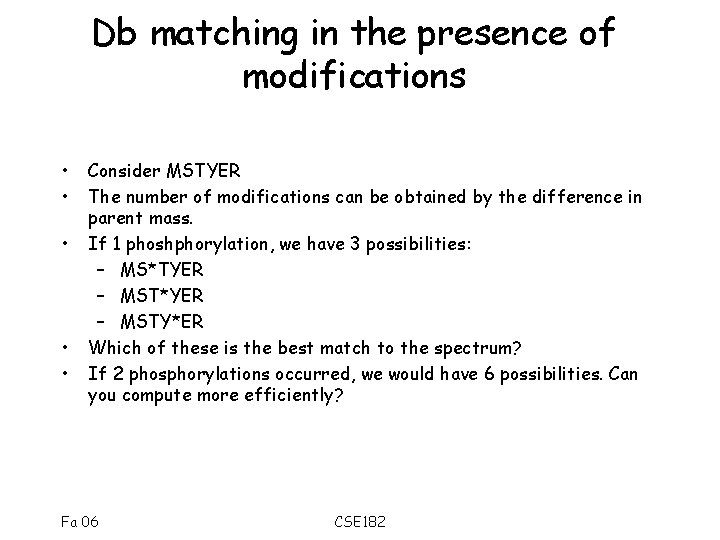
Db matching in the presence of modifications • • • Consider MSTYER The number of modifications can be obtained by the difference in parent mass. If 1 phoshphorylation, we have 3 possibilities: – MS*TYER – MST*YER – MSTY*ER Which of these is the best match to the spectrum? If 2 phosphorylations occurred, we would have 6 possibilities. Can you compute more efficiently? Fa 06 CSE 182

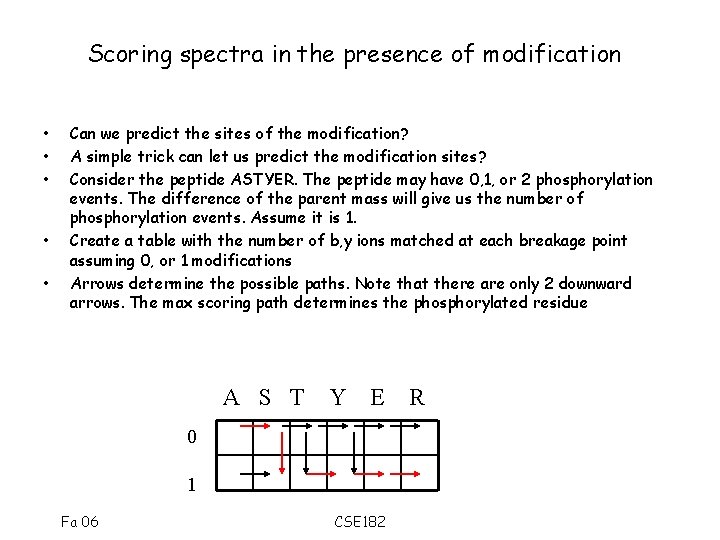
Scoring spectra in the presence of modification • • • Can we predict the sites of the modification? A simple trick can let us predict the modification sites? Consider the peptide ASTYER. The peptide may have 0, 1, or 2 phosphorylation events. The difference of the parent mass will give us the number of phosphorylation events. Assume it is 1. Create a table with the number of b, y ions matched at each breakage point assuming 0, or 1 modifications Arrows determine the possible paths. Note that there are only 2 downward arrows. The max scoring path determines the phosphorylated residue A S T Y E 0 1 Fa 06 CSE 182 R

Modifications • Modifications significantly increase the time of search. • The algorithm speeds it up somewhat, but is still expensive Fa 06 CSE 182

Fast identification of modified peptides Fa 06 CSE 182

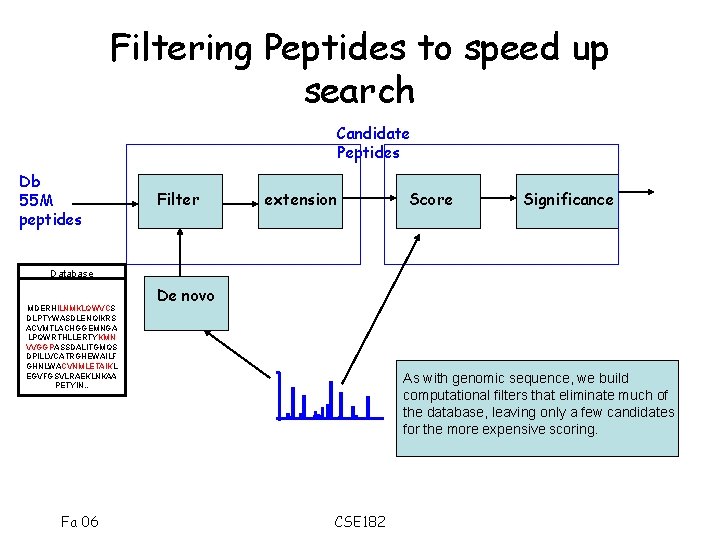
Filtering Peptides to speed up search Candidate Peptides Db 55 M peptides Filter extension Score Significance Database MDERHILNMKLQWVCS DLPTYWASDLENQIKRS ACVMTLACHGGEMNGA LPQWRTHLLERTYKMN VVGGPASSDALITGMQS DPILLVCATRGHEWAILF GHNLWACVNMLETAIKL EGVFGSVLRAEKLNKAA PETYIN. . Fa 06 De novo As with genomic sequence, we build computational filters that eliminate much of the database, leaving only a few candidates for the more expensive scoring. CSE 182

Basic Filtering • Typical tools score all peptides with close enough parent mass and tryptic termini • Filtering by parent mass is problematic when PTMs are allowed, as one must consider multiple parent masses Fa 06 CSE 182

Tag-based filtering • A tag is a short peptide with a prefix and suffix mass • Efficient: An average tripeptide tag matches Swiss-Prot ~700 times • Analogy: Using tags to search the proteome is similar to moving from full Smith-Waterman alignment to BLAST Fa 06 CSE 182

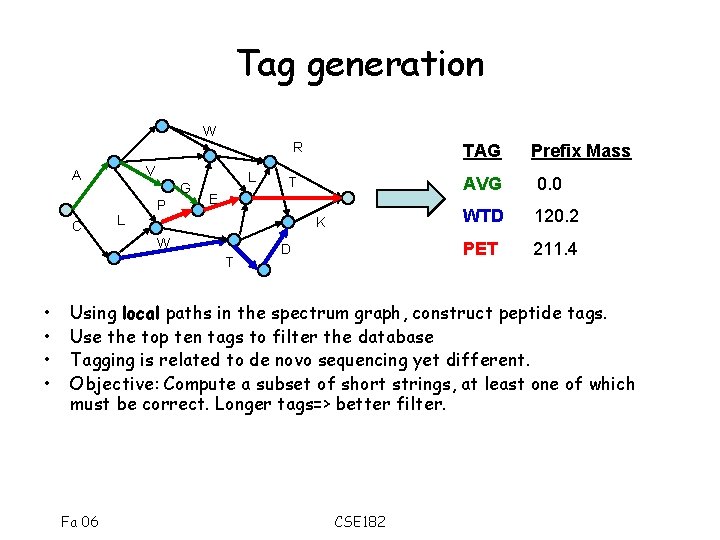
Tag generation W V A C R G L P L T E K W T • • TAG D Prefix Mass AVG 0. 0 WTD 120. 2 PET 211. 4 Using local paths in the spectrum graph, construct peptide tags. Use the top ten tags to filter the database Tagging is related to de novo sequencing yet different. Objective: Compute a subset of short strings, at least one of which must be correct. Longer tags=> better filter. Fa 06 CSE 182

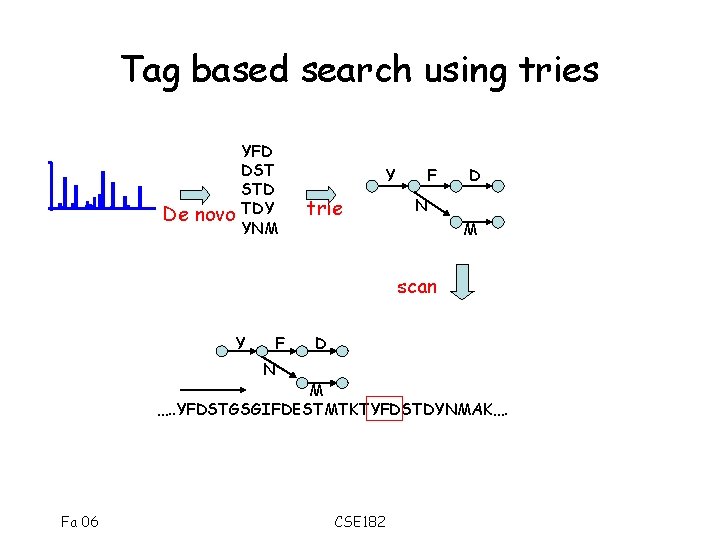
Tag based search using tries De novo YFD DST STD TDY YNM Y trie F D N M scan Y F D N M …. . YFDSTGSGIFDESTMTKTYFDSTDYNMAK…. Fa 06 CSE 182

Modification Summary • Modifications shift spectra in characteristic ways. • A modification sensitive database search can identify modifications, but is computationally expensive • Filtering using de novo tag generation can speed up the process making identification of modified peptides tractable. Fa 06 CSE 182

MS based quantitation Fa 06 CSE 182

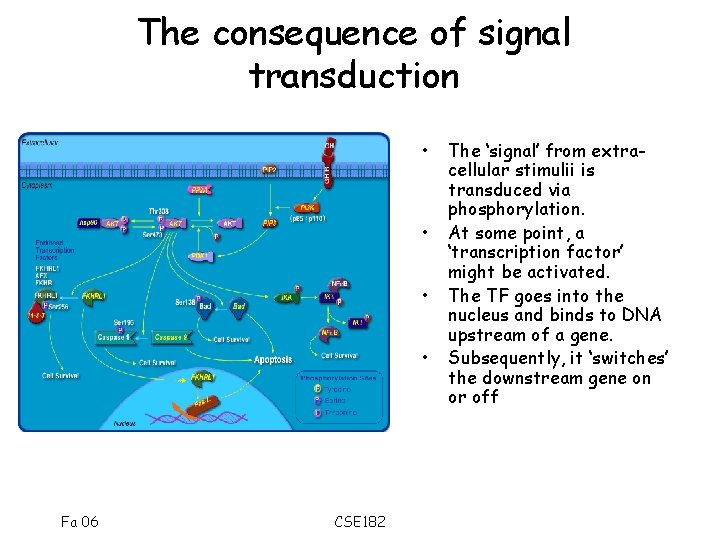
The consequence of signal transduction • • Fa 06 CSE 182 The ‘signal’ from extracellular stimulii is transduced via phosphorylation. At some point, a ‘transcription factor’ might be activated. The TF goes into the nucleus and binds to DNA upstream of a gene. Subsequently, it ‘switches’ the downstream gene on or off

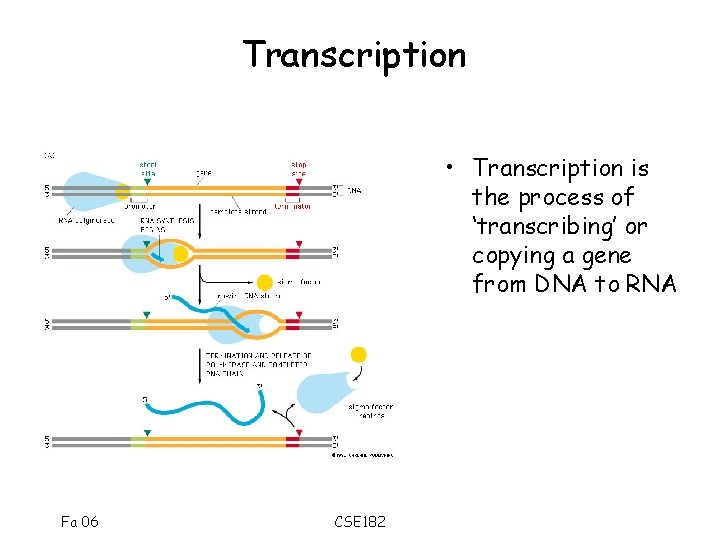
Transcription • Transcription is the process of ‘transcribing’ or copying a gene from DNA to RNA Fa 06 CSE 182

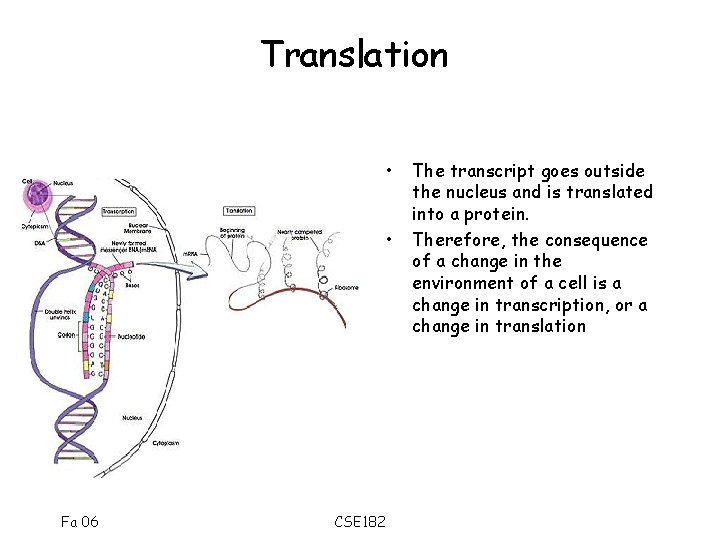
Translation • • Fa 06 CSE 182 The transcript goes outside the nucleus and is translated into a protein. Therefore, the consequence of a change in the environment of a cell is a change in transcription, or a change in translation

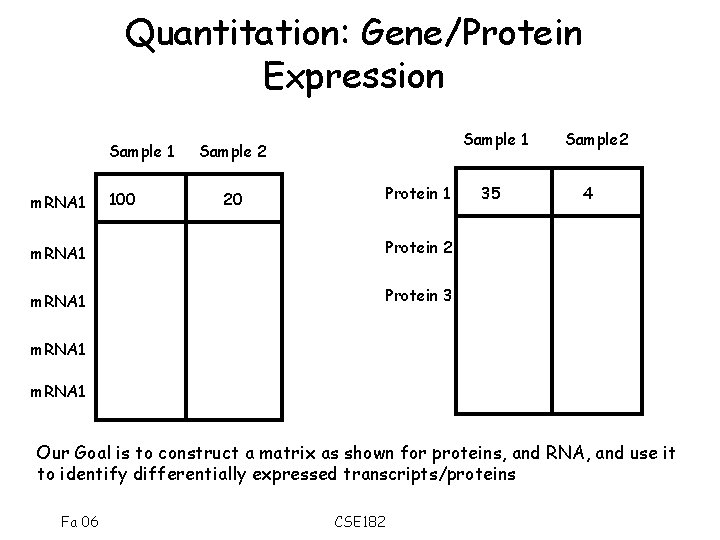
Quantitation: Gene/Protein Expression Sample 1 m. RNA 1 100 Sample 1 Sample 2 Protein 1 20 m. RNA 1 Protein 2 m. RNA 1 Protein 3 35 Sample 2 4 m. RNA 1 Our Goal is to construct a matrix as shown for proteins, and RNA, and use it to identify differentially expressed transcripts/proteins Fa 06 CSE 182

Gene Expression • Measuring expression at transcript level is done by microarrays and other tools • Expression at the protein level is being done using mass spectrometry. • Two problems arise: – Data: How to populate the matrices on the previous slide? (‘easy’ for m. RNA, difficult for proteins) – Analysis: Is a change in expression significant? (Identical for both m. RNA, and proteins). • We will consider the data problem here. The analysis problem will be considered when we discuss micro-arrays. Fa 06 CSE 182
 Cse 182
Cse 182 Cse 182
Cse 182 Cse 182 ucsd
Cse 182 ucsd Peptide identification
Peptide identification Peptide identification
Peptide identification Mass spectrometry a level
Mass spectrometry a level Mass spectrometry
Mass spectrometry Mass spectrometer block diagram
Mass spectrometer block diagram Khan academy mass spectrometry
Khan academy mass spectrometry Mass spectrometry
Mass spectrometry Mass spectrometry in forensic science
Mass spectrometry in forensic science Sympatry
Sympatry Dating
Dating How to read mass spectrometry graph
How to read mass spectrometry graph Mass spectrometry
Mass spectrometry Mass spectrometry exam questions
Mass spectrometry exam questions Mass spectrometry ionization
Mass spectrometry ionization Nitrogen rule in mass spectrometry
Nitrogen rule in mass spectrometry Swath mass spectrometry
Swath mass spectrometry Sdbsweb
Sdbsweb Mass spectrometry problem set
Mass spectrometry problem set Components of mass spectrometer
Components of mass spectrometer Mass spectrometry
Mass spectrometry Mass spectrometry lecture
Mass spectrometry lecture Mass spectrometer block diagram
Mass spectrometer block diagram Mass spectrometry
Mass spectrometry Mass spectrometry data acquisition for gc/ms
Mass spectrometry data acquisition for gc/ms Deflection in mass spectrometry
Deflection in mass spectrometry Inlet system in mass spectrometry
Inlet system in mass spectrometry Chromosomes and alleles
Chromosomes and alleles Mascot peptide mass fingerprint
Mascot peptide mass fingerprint Central pocket whorl vs plain whorl
Central pocket whorl vs plain whorl Department order 182
Department order 182 Cessna 182 diesel
Cessna 182 diesel Heer ons dink aan afrika
Heer ons dink aan afrika Cs 182
Cs 182 Tungsten-182
Tungsten-182 Engine on half power ahead - steer 182 degrees port side.
Engine on half power ahead - steer 182 degrees port side. Love 182
Love 182 Nosotros commands
Nosotros commands Min 167 + 182
Min 167 + 182 Berkeley cs 182
Berkeley cs 182 Cold vapor atomic fluorescence spectrometry
Cold vapor atomic fluorescence spectrometry Cold vapor atomic fluorescence spectrometry
Cold vapor atomic fluorescence spectrometry