Milk a source of nourishment for all mammals



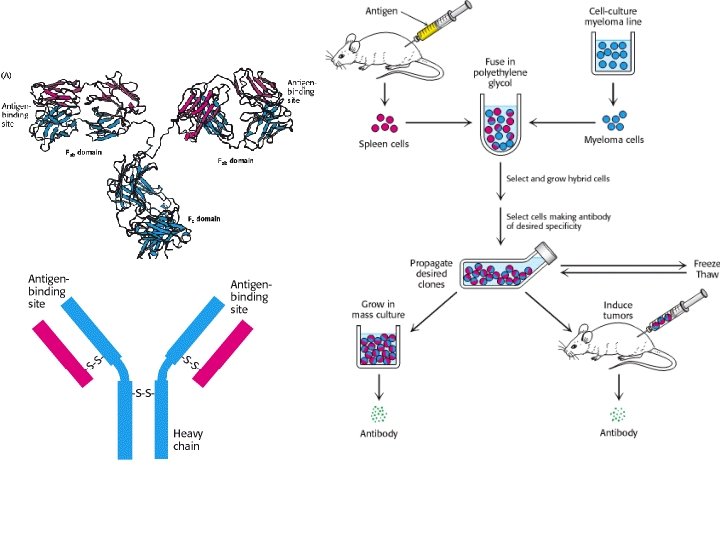





- Slides: 25

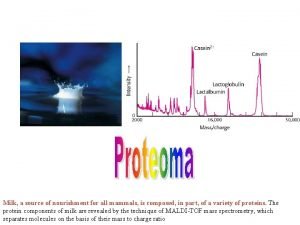
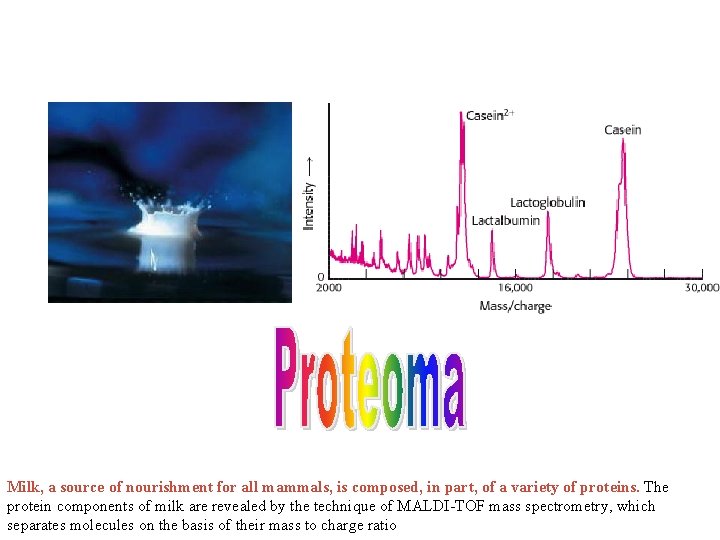
Milk, a source of nourishment for all mammals, is composed, in part, of a variety of proteins. The protein components of milk are revealed by the technique of MALDI-TOF mass spectrometry, which separates molecules on the basis of their mass to charge ratio

Liberar a proteína da célula para purificá-la Figure 4. 1. Differential Centrifugation. Cells are disrupted in a homogenizer and the resulting mixture, called the homogenate, is centrifuged in a step-by-step fashion of increasing centrifugal force. The denser material will form a pellet at lower centrifugal force than will the less-dense material. The isolated fractions can be used for further purification.

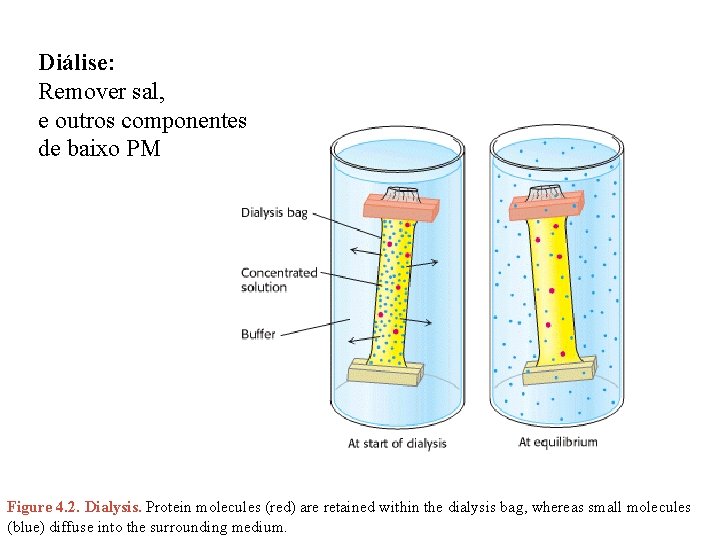
Diálise: Remover sal, e outros componentes de baixo PM Figure 4. 2. Dialysis. Protein molecules (red) are retained within the dialysis bag, whereas small molecules (blue) diffuse into the surrounding medium.

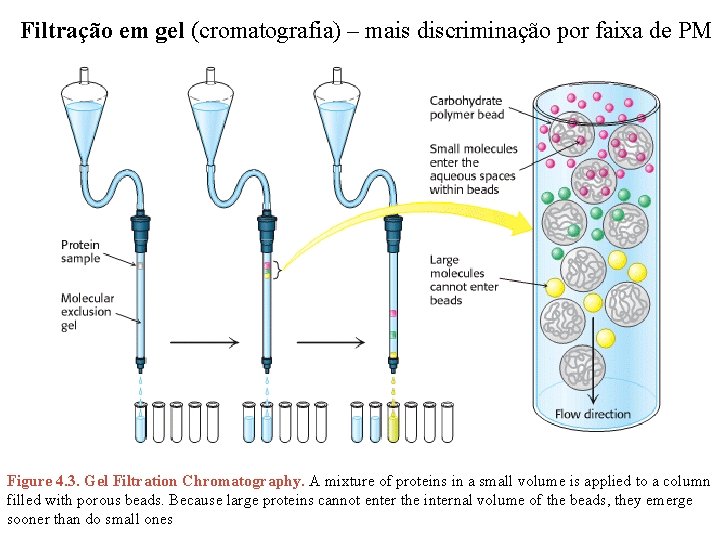
Filtração em gel (cromatografia) – mais discriminação por faixa de PM Figure 4. 3. Gel Filtration Chromatography. A mixture of proteins in a small volume is applied to a column filled with porous beads. Because large proteins cannot enter the internal volume of the beads, they emerge sooner than do small ones

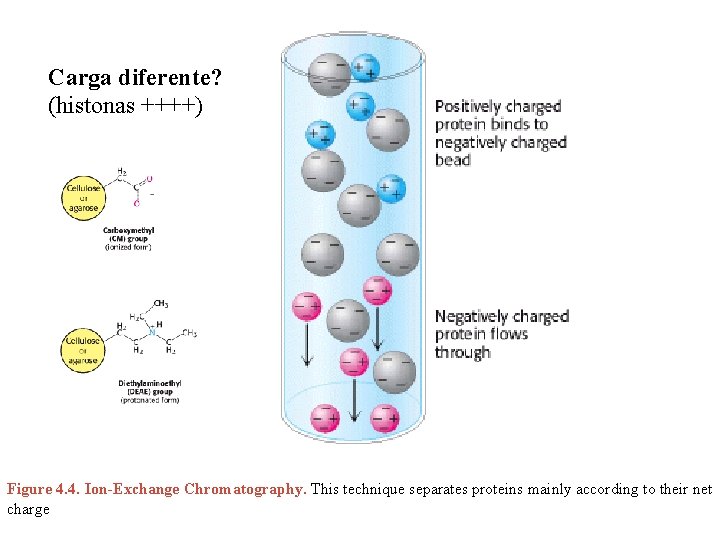
Carga diferente? (histonas ++++) Figure 4. 4. Ion-Exchange Chromatography. This technique separates proteins mainly according to their net charge

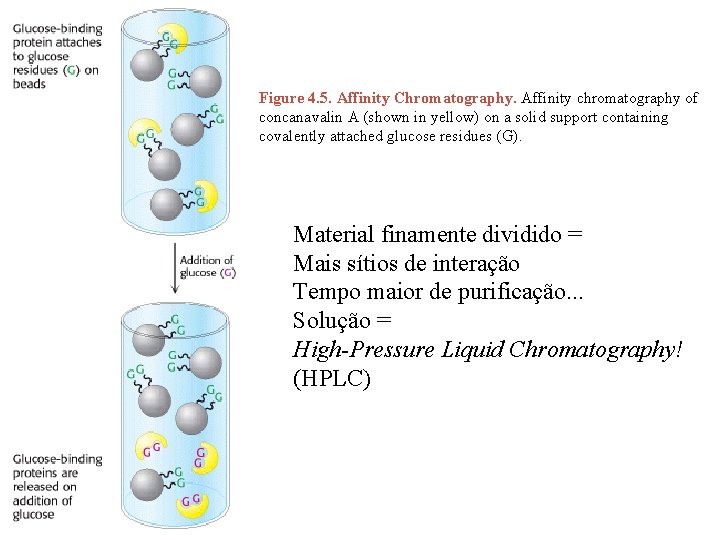
Figure 4. 5. Affinity Chromatography. Affinity chromatography of concanavalin A (shown in yellow) on a solid support containing covalently attached glucose residues (G). Material finamente dividido = Mais sítios de interação Tempo maior de purificação. . . Solução = High-Pressure Liquid Chromatography! (HPLC)

EXEMPLO de HPLC Figure 4. 6. High-Pressure Liquid Chromatography (HPLC). Gel filtration by HPLC clearly defines the individual proteins because of its greater resolving power: (1) thyroglobulin (669 kd), (2) catalase (232 kd), (3) bovine serum albumin (67 kd), (4) ovalbumin (43 kd), and (5) ribonuclease (13. 4 kd).

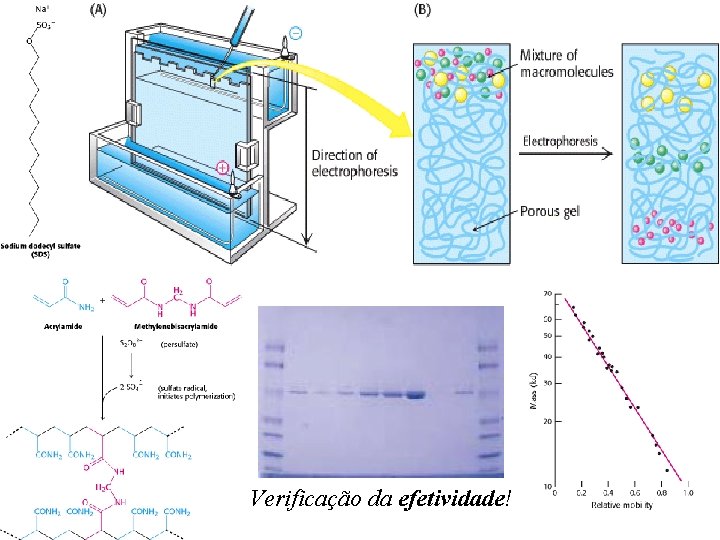
Verificação da efetividade!

Composição de AA: primeira etapa de seqüenciamento Figure 4. 18. Determination of Amino Acid Composition. Different amino acids in a peptide hydrolysate can be separated by ion-exchange chromatography on a sulfonated polystyrene resin (such as Dowex-50). Buffers (in this case, sodium citrate) of increasing p. H are used to elute the amino acids from the column. The amount of each amino acid present is determined from the absorbance. Aspartate, which has an acidic side chain, is first to emerge, whereas arginine, which has a basic side chain, is the last. The original peptide is revealed to be composed of one aspartate, one alanine, one phenylalanine, one arginine, and two glycine residues

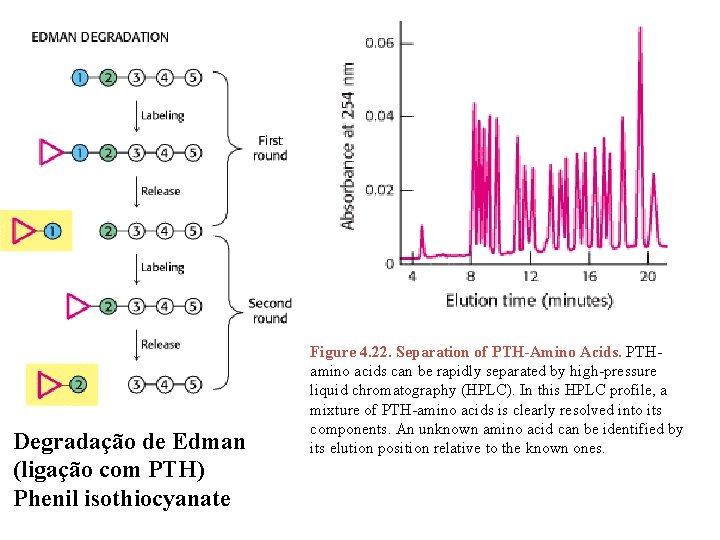
Degradação de Edman (ligação com PTH) Phenil isothiocyanate Figure 4. 22. Separation of PTH-Amino Acids. PTHamino acids can be rapidly separated by high-pressure liquid chromatography (HPLC). In this HPLC profile, a mixture of PTH-amino acids is clearly resolved into its components. An unknown amino acid can be identified by its elution position relative to the known ones.

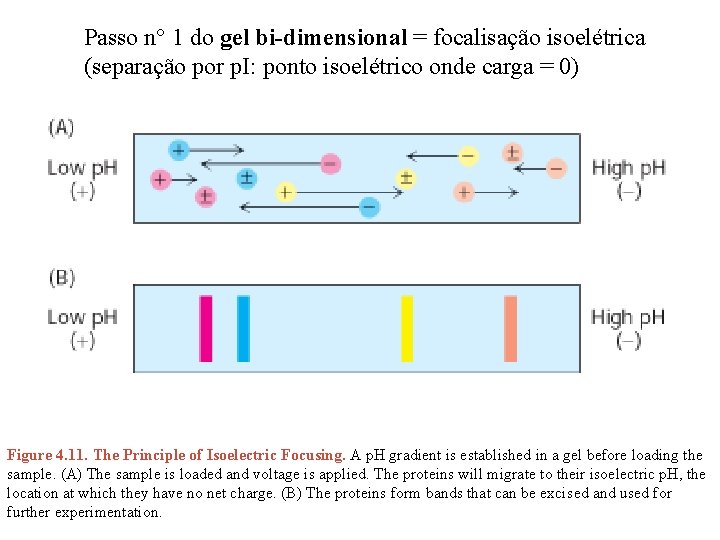
Passo n° 1 do gel bi-dimensional = focalisação isoelétrica (separação por p. I: ponto isoelétrico onde carga = 0) Figure 4. 11. The Principle of Isoelectric Focusing. A p. H gradient is established in a gel before loading the sample. (A) The sample is loaded and voltage is applied. The proteins will migrate to their isoelectric p. H, the location at which they have no net charge. (B) The proteins form bands that can be excised and used for further experimentation.

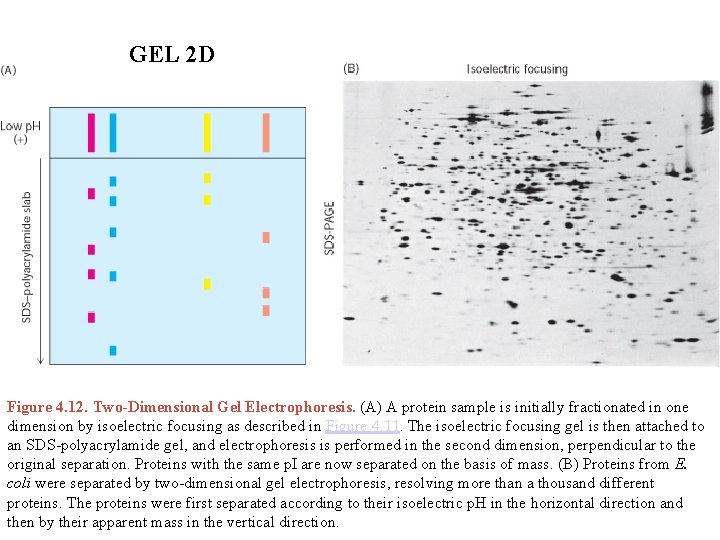
GEL 2 D Figure 4. 12. Two-Dimensional Gel Electrophoresis. (A) A protein sample is initially fractionated in one dimension by isoelectric focusing as described in Figure 4. 11. The isoelectric focusing gel is then attached to an SDS-polyacrylamide gel, and electrophoresis is performed in the second dimension, perpendicular to the original separation. Proteins with the same p. I are now separated on the basis of mass. (B) Proteins from E. coli were separated by two-dimensional gel electrophoresis, resolving more than a thousand different proteins. The proteins were first separated according to their isoelectric p. H in the horizontal direction and then by their apparent mass in the vertical direction.

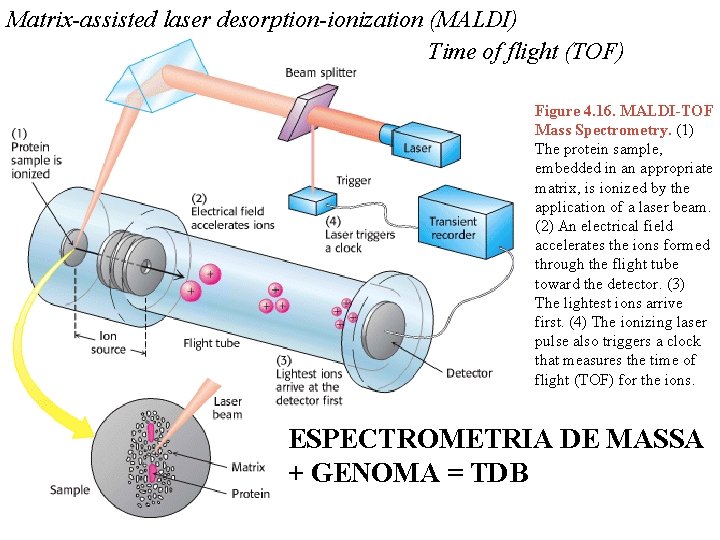
Matrix-assisted laser desorption-ionization (MALDI) Time of flight (TOF) Figure 4. 16. MALDI-TOF Mass Spectrometry. (1) The protein sample, embedded in an appropriate matrix, is ionized by the application of a laser beam. (2) An electrical field accelerates the ions formed through the flight tube toward the detector. (3) The lightest ions arrive first. (4) The ionizing laser pulse also triggers a clock that measures the time of flight (TOF) for the ions. ESPECTROMETRIA DE MASSA + GENOMA = TDB

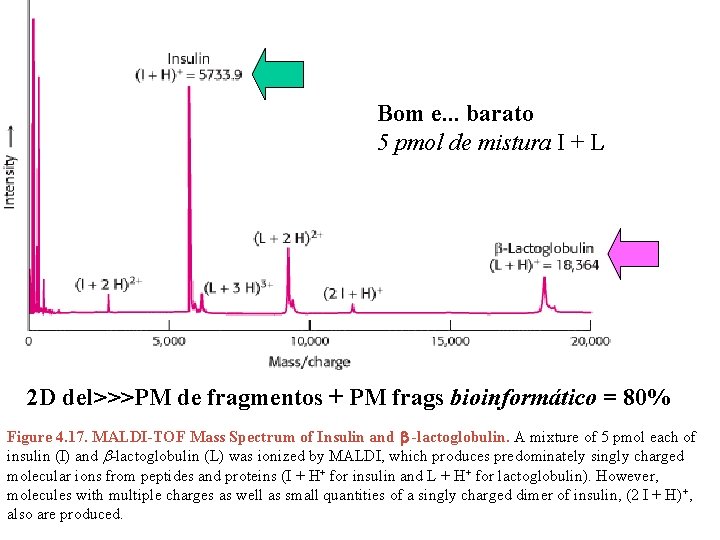
Bom e. . . barato 5 pmol de mistura I + L 2 D del>>>PM de fragmentos + PM frags bioinformático = 80% Figure 4. 17. MALDI-TOF Mass Spectrum of Insulin and b -lactoglobulin. A mixture of 5 pmol each of insulin (I) and b-lactoglobulin (L) was ionized by MALDI, which produces predominately singly charged molecular ions from peptides and proteins (I + H+ for insulin and L + H+ for lactoglobulin). However, molecules with multiple charges as well as small quantities of a singly charged dimer of insulin, (2 I + H)+, also are produced.


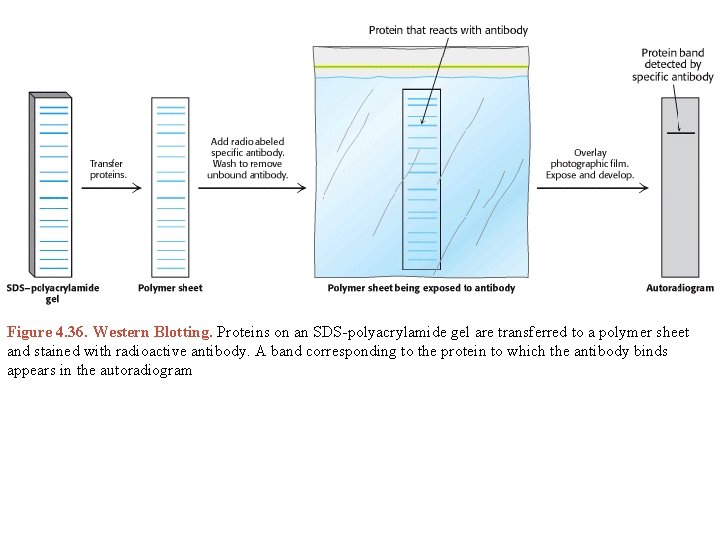
Figure 4. 36. Western Blotting. Proteins on an SDS-polyacrylamide gel are transferred to a polymer sheet and stained with radioactive antibody. A band corresponding to the protein to which the antibody binds appears in the autoradiogram

Figure 4. 35. Indirect ELISA and Sandwich ELISA (A) In indirect ELISA, the production of color indicates the amount of an antibody to a specific antigen. (B) In sandwich ELISA, the production of color indicates the quantity of antigen

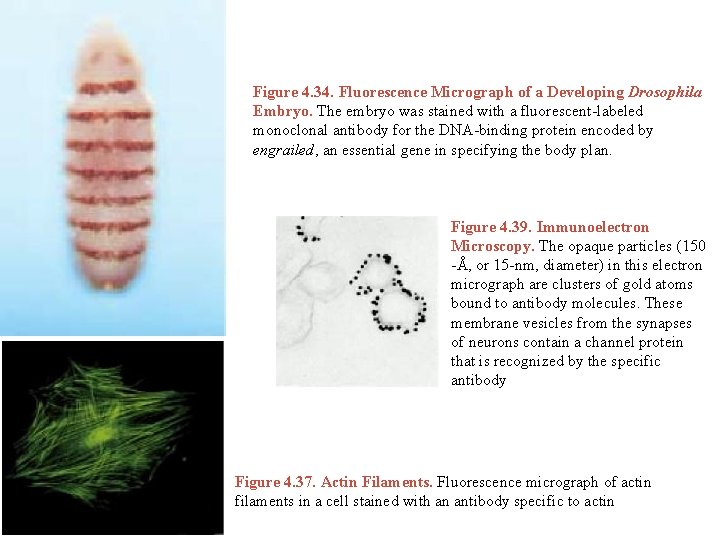
Figure 4. 34. Fluorescence Micrograph of a Developing Drosophila Embryo. The embryo was stained with a fluorescent-labeled monoclonal antibody for the DNA-binding protein encoded by engrailed, an essential gene in specifying the body plan. Figure 4. 39. Immunoelectron Microscopy. The opaque particles (150 -Å, or 15 -nm, diameter) in this electron micrograph are clusters of gold atoms bound to antibody molecules. These membrane vesicles from the synapses of neurons contain a channel protein that is recognized by the specific antibody Figure 4. 37. Actin Filaments. Fluorescence micrograph of actin filaments in a cell stained with an antibody specific to actin

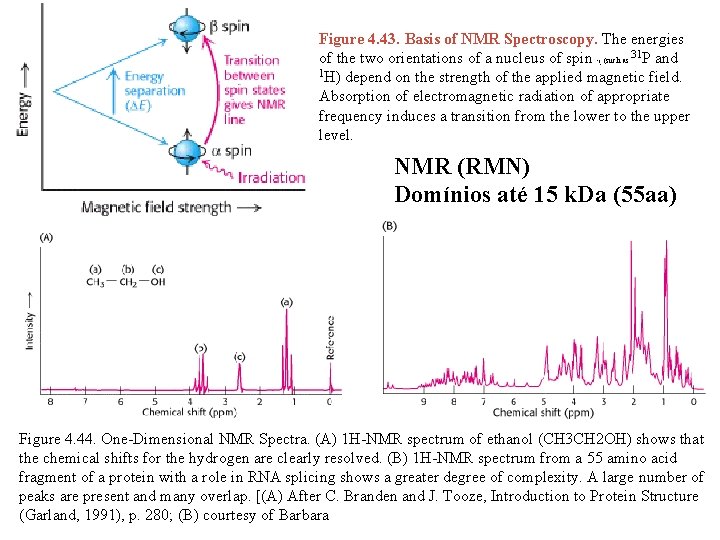
Figure 4. 43. Basis of NMR Spectroscopy. The energies of the two orientations of a nucleus of spin (such as 31 P and 1 H) depend on the strength of the applied magnetic field. Absorption of electromagnetic radiation of appropriate frequency induces a transition from the lower to the upper level. 1/ 2 NMR (RMN) Domínios até 15 k. Da (55 aa) Figure 4. 44. One-Dimensional NMR Spectra. (A) 1 H-NMR spectrum of ethanol (CH 3 CH 2 OH) shows that the chemical shifts for the hydrogen are clearly resolved. (B) 1 H-NMR spectrum from a 55 amino acid fragment of a protein with a role in RNA splicing shows a greater degree of complexity. A large number of peaks are present and many overlap. [(A) After C. Branden and J. Tooze, Introduction to Protein Structure (Garland, 1991), p. 280; (B) courtesy of Barbara

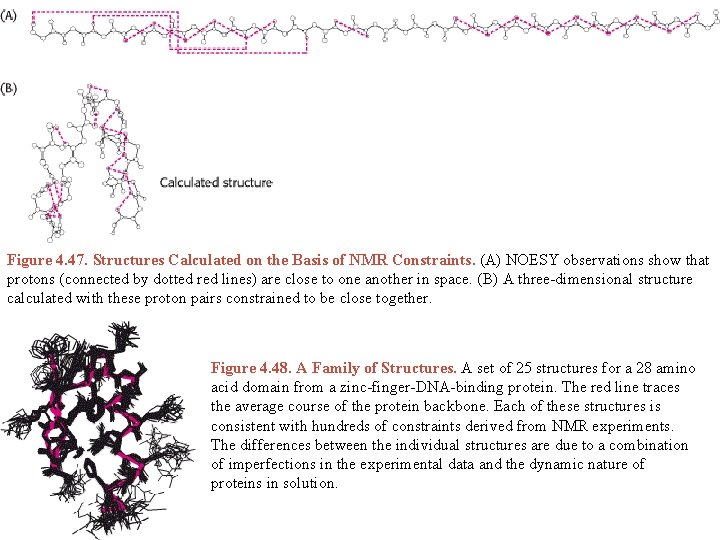
Figure 4. 47. Structures Calculated on the Basis of NMR Constraints. (A) NOESY observations show that protons (connected by dotted red lines) are close to one another in space. (B) A three-dimensional structure calculated with these proton pairs constrained to be close together. Figure 4. 48. A Family of Structures. A set of 25 structures for a 28 amino acid domain from a zinc-finger-DNA-binding protein. The red line traces the average course of the protein backbone. Each of these structures is consistent with hundreds of constraints derived from NMR experiments. The differences between the individual structures are due to a combination of imperfections in the experimental data and the dynamic nature of proteins in solution.

Figure 4. 49. Essence of an X-Ray Crystallographic Experiment: an X-Ray Beam, a Crystal, and a Detector.

Figure 4. 51. Myoglobin Crystal and X-Ray. (A) Crystal of myoglobin. (B) X-ray precession photograph of a myoglobin crystal. Figure 4. 52. Section of the Electron-Density Map of Myoglobin. This section of the electron-density map shows the heme group. The peak of the center of this section corresponds to the position of the iron atom.



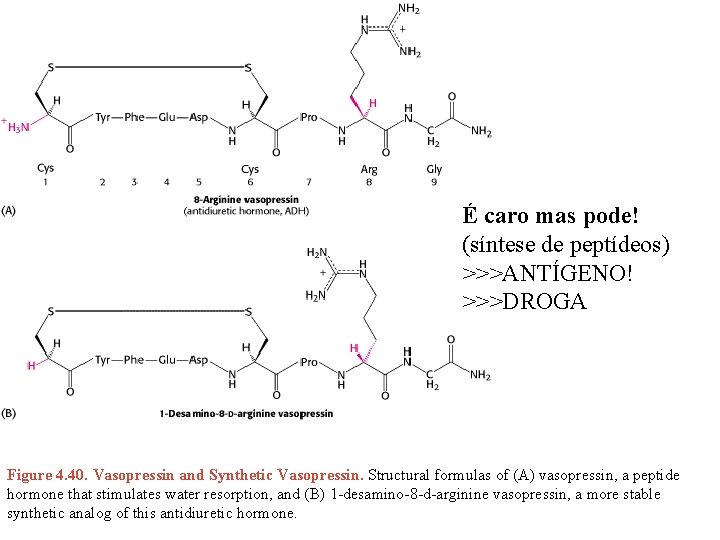
É caro mas pode! (síntese de peptídeos) >>>ANTÍGENO! >>>DROGA Figure 4. 40. Vasopressin and Synthetic Vasopressin. Structural formulas of (A) vasopressin, a peptide hormone that stimulates water resorption, and (B) 1 -desamino-8 -d-arginine vasopressin, a more stable synthetic analog of this antidiuretic hormone.
 Pelycosaurs
Pelycosaurs Fish classification of vertebrates
Fish classification of vertebrates Food web
Food web Chapparal food web
Chapparal food web Sensory evaluation of milk and milk products
Sensory evaluation of milk and milk products Milk for toddlers with milk allergynon dairy
Milk for toddlers with milk allergynon dairy Human milk vs cow milk
Human milk vs cow milk Contamination of milk and milk products
Contamination of milk and milk products G
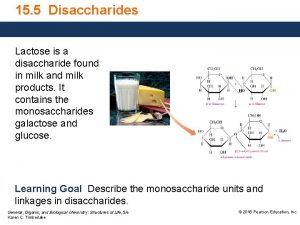
G Disaccharide found in milk and milk products
Disaccharide found in milk and milk products What is a mammal
What is a mammal Three basic scale patterns hair
Three basic scale patterns hair All mammals have hair. its main purpose is to
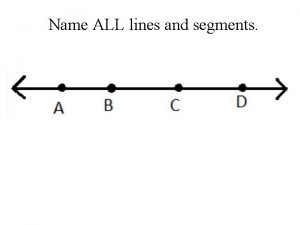
All mammals have hair. its main purpose is to Name a point that is collinear with the given points
Name a point that is collinear with the given points Fspos
Fspos Typiska drag för en novell
Typiska drag för en novell Tack för att ni lyssnade bild
Tack för att ni lyssnade bild Returpilarna
Returpilarna Varför kallas perioden 1918-1939 för mellankrigstiden?
Varför kallas perioden 1918-1939 för mellankrigstiden? En lathund för arbete med kontinuitetshantering
En lathund för arbete med kontinuitetshantering Kassaregister ideell förening
Kassaregister ideell förening Personlig tidbok för yrkesförare
Personlig tidbok för yrkesförare Sura för anatom
Sura för anatom Vad är densitet
Vad är densitet Datorkunskap för nybörjare
Datorkunskap för nybörjare Stig kerman
Stig kerman