Data Acquisition and Analysis in Mass Spectrometry Based

Data Acquisition and Analysis in Mass Spectrometry Based Metabolomics Pavel Aronov Bio. Cyc workshop October 27, 2010

Outline n n n Fundamentals of Mass Spectrometry Data Acquisition and Analysis in GCMS based Metabolomics Data Acquisition and Analysis in LCMS based Metabolomics
How to analyze tryptophan or any other metabolite? Two most common techniques in analytical chemistry to determine or confirm chemical structure: n Nuclear Magnetic Resonance/NMR (1940 s, Felix Bloch at Stanford University) Excellent structural information n Mass Spectrometry (1900 s, JJ Thompson at Cambridge University) Excellent sensitivity
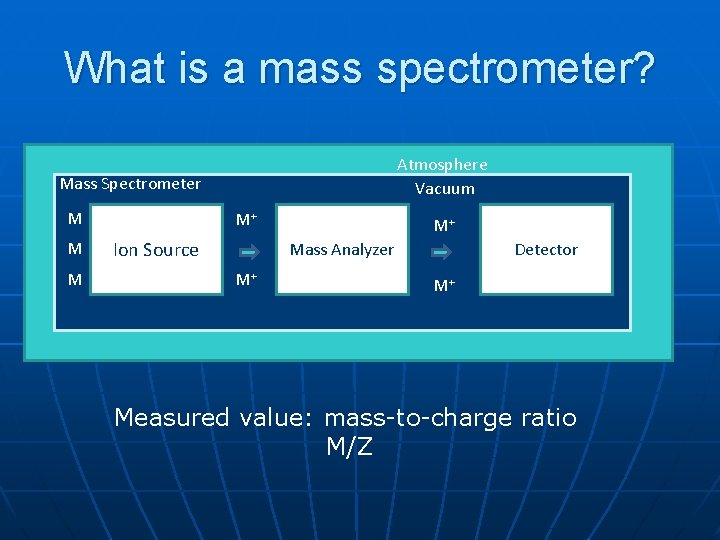
What is a mass spectrometer? Atmosphere Vacuum Mass Spectrometer M M+ Ion Source M+ Mass Analyzer M+ Detector M+ Measured value: mass-to-charge ratio M/Z

Mass Units n Unit of mass: 1/12 mass of carbon-12 atom 1 u or 1 Da n Unit of mass-to-charge 1 Da / z = 1 Th (Thompson) m/z 205 For metabolites usually z = 1, Hence 1 Da is equivalent to 1 Th
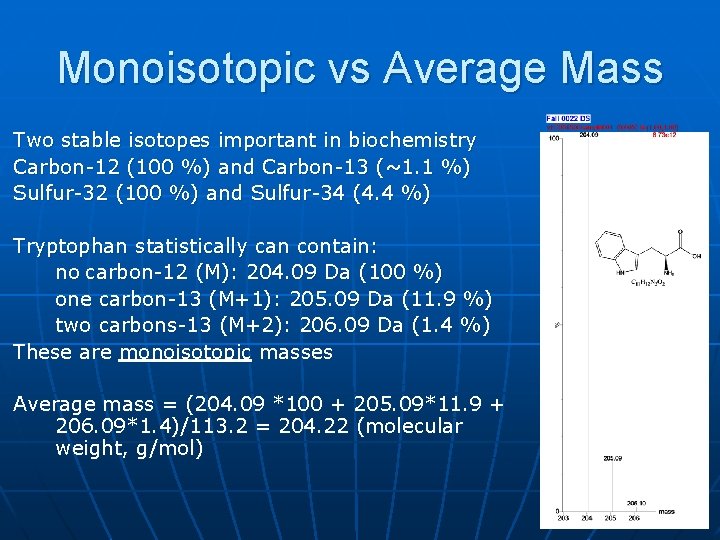
Monoisotopic vs Average Mass Two stable isotopes important in biochemistry Carbon-12 (100 %) and Carbon-13 (~1. 1 %) Sulfur-32 (100 %) and Sulfur-34 (4. 4 %) Tryptophan statistically can contain: no carbon-12 (M): 204. 09 Da (100 %) one carbon-13 (M+1): 205. 09 Da (11. 9 %) two carbons-13 (M+2): 206. 09 Da (1. 4 %) These are monoisotopic masses Average mass = (204. 09 *100 + 205. 09*11. 9 + 206. 09*1. 4)/113. 2 = 204. 22 (molecular weight, g/mol)

Mass defect 1 H (p+e-) 12 C 14 N 16 O n 1. 0078 u 12. 0000 u 14. 0031 u 15. 9949 u 1. 0087 u Carbon-12: 6 protons, 6 neutron and 6 electrons 6 x 1. 0078 u + 6 x 1. 0087 u = 12. 0990 u Mass Defect = 12. 0990 u – 12. 0000 u = 0. 0990 u E = mc 2 0. 1 u = 93 Me. V
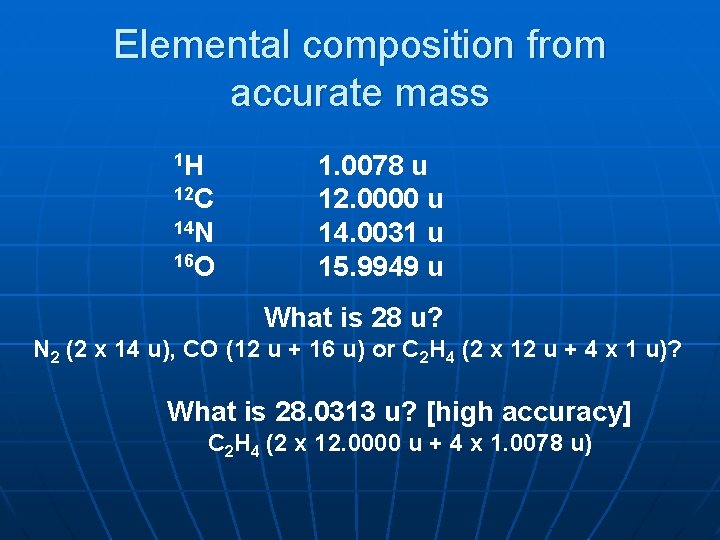
Elemental composition from accurate mass 1 H 12 C 14 N 16 O 1. 0078 u 12. 0000 u 14. 0031 u 15. 9949 u What is 28 u? N 2 (2 x 14 u), CO (12 u + 16 u) or C 2 H 4 (2 x 12 u + 4 x 1 u)? What is 28. 0313 u? [high accuracy] C 2 H 4 (2 x 12. 0000 u + 4 x 1. 0078 u)
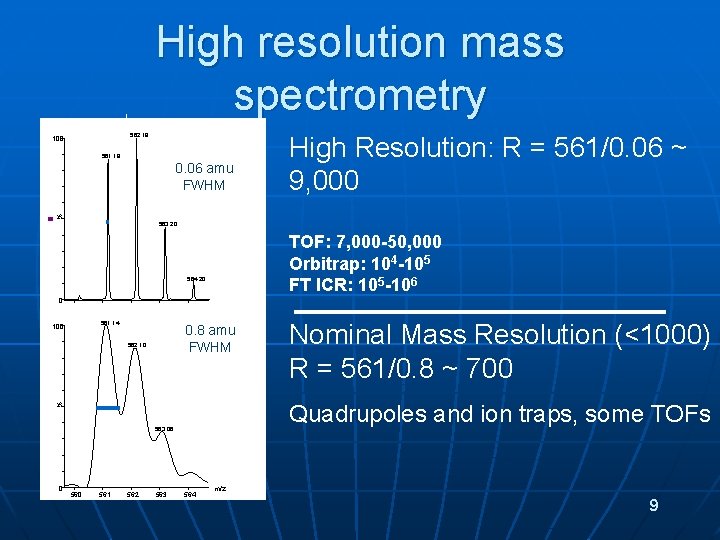
High resolution mass spectrometry 562. 19 100 561. 18 % 0. 06 amu FWHM High Resolution: R = 561/0. 06 ~ 9, 000 563. 20 TOF: 7, 000 -50, 000 Orbitrap: 104 -105 FT ICR: 105 -106 564. 20 0 561. 14 100 0. 8 amu FWHM 563. 06 0 561 562 563 Nominal Mass Resolution (<1000) R = 561/0. 8 ~ 700 Quadrupoles and ion traps, some TOFs % 562. 10 564 m/z 9
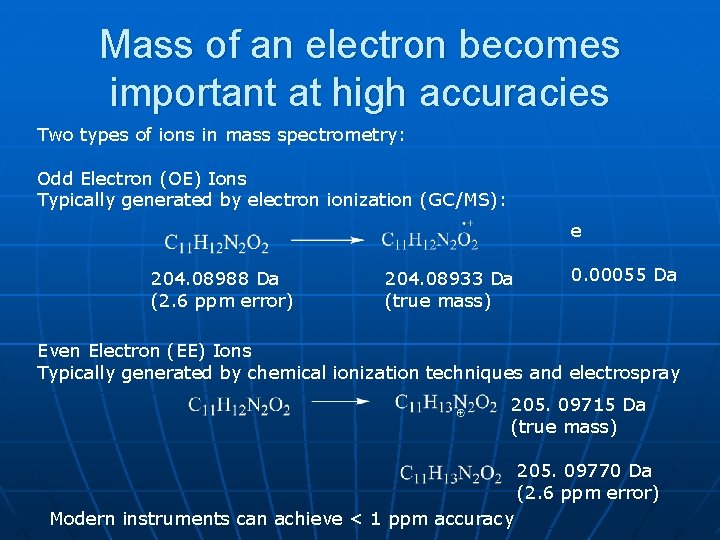
Mass of an electron becomes important at high accuracies Two types of ions in mass spectrometry: Odd Electron (OE) Ions Typically generated by electron ionization (GC/MS): e 204. 08988 Da (2. 6 ppm error) 204. 08933 Da (true mass) 0. 00055 Da Even Electron (EE) Ions Typically generated by chemical ionization techniques and electrospray 205. 09715 Da (true mass) 205. 09770 Da (2. 6 ppm error) Modern instruments can achieve < 1 ppm accuracy
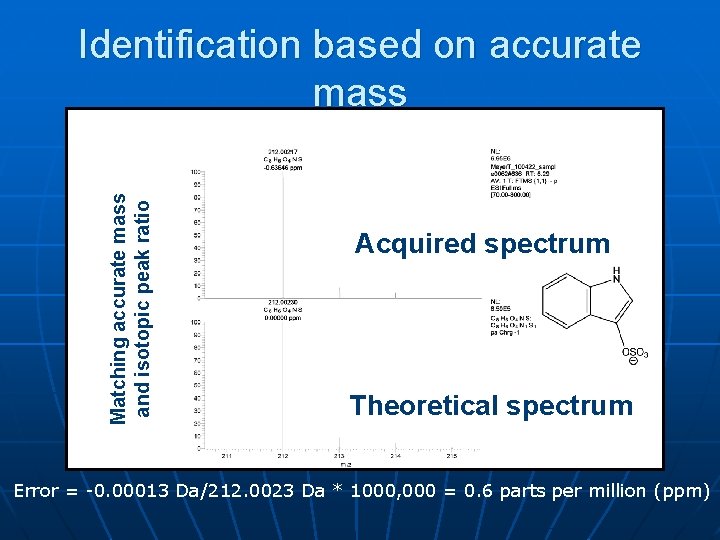
Matching accurate mass and isotopic peak ratio Identification based on accurate mass Acquired spectrum Theoretical spectrum Error = -0. 00013 Da/212. 0023 Da * 1000, 000 = 0. 6 parts per million (ppm)
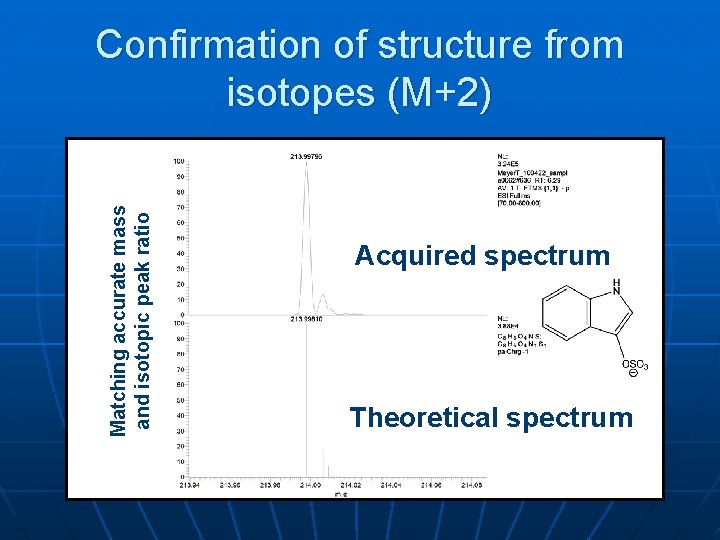
Matching accurate mass and isotopic peak ratio Confirmation of structure from isotopes (M+2) Acquired spectrum Theoretical spectrum

Tandem Mass Spectrometry Mass Spectrometer M M M Ion Source HPLC M+ Mass M+ Analyzer M+ 1 M+ Collision Cell Atmosphere Vacuum F+ F+ Mass Analyzer 2 F+ Detector F+
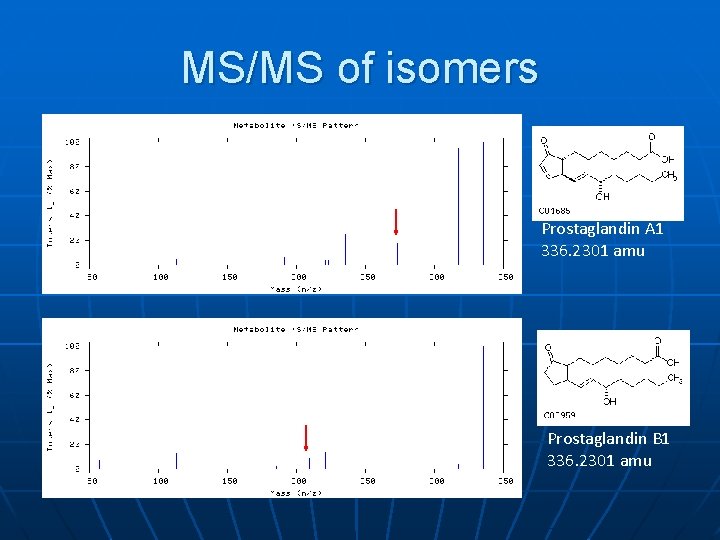
MS/MS of isomers Prostaglandin A 1 336. 2301 amu Prostaglandin B 1 336. 2301 amu
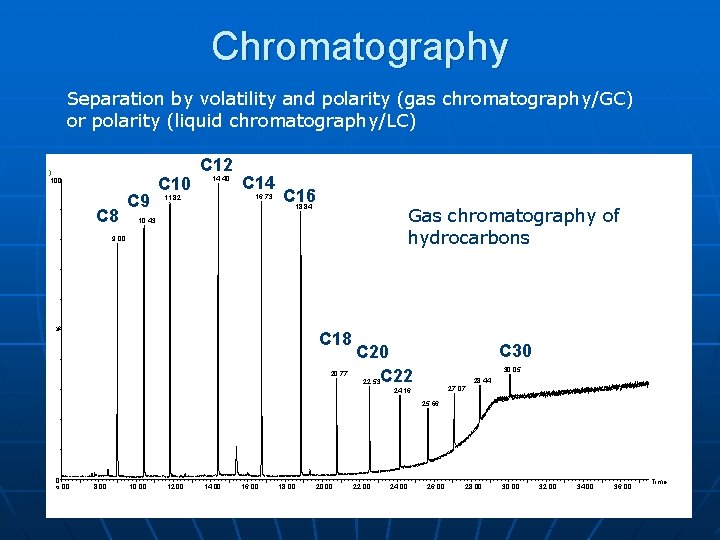
Chromatography Separation by volatility and polarity (gas chromatography/GC) or polarity (liquid chromatography/LC) ) 100 C 8 C 9 C 10 C 12 14. 40 C 14 16. 73 11. 82 C 16 18. 84 Gas chromatography of hydrocarbons 10. 43 % 9. 00 C 18 20. 77 C 30 C 22 30. 05 28. 44 22. 53 27. 07 24. 16 25. 66 0 6. 00 8. 00 10. 00 12. 00 14. 00 16. 00 18. 00 20. 00 22. 00 24. 00 26. 00 28. 00 30. 00 32. 00 34. 00 36. 00 Time
2 D dimensionality of metabolomics data in LC-MS and GC-MS
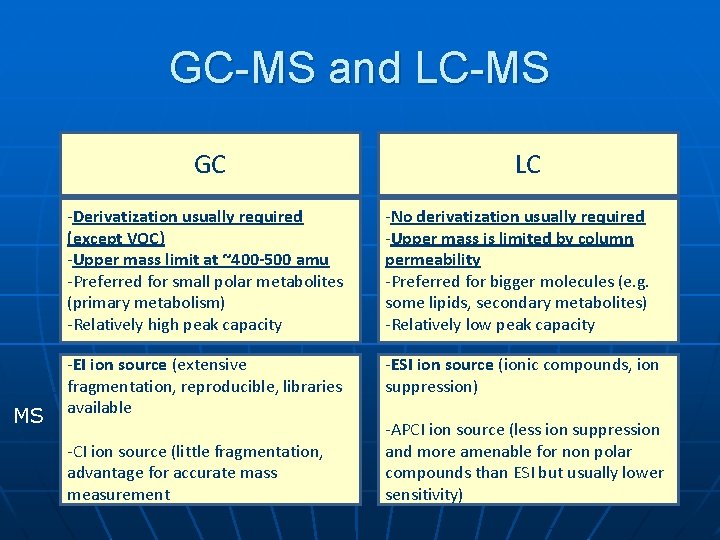
GC-MS and LC-MS GC MS LC -Derivatization usually required (except VOC) -Upper mass limit at ~400 -500 amu -Preferred for small polar metabolites (primary metabolism) -Relatively high peak capacity -No derivatization usually required -Upper mass is limited by column permeability -Preferred for bigger molecules (e. g. some lipids, secondary metabolites) -Relatively low peak capacity -EI ion source (extensive fragmentation, reproducible, libraries available -ESI ion source (ionic compounds, ion suppression) -CI ion source (little fragmentation, advantage for accurate mass measurement -APCI ion source (less ion suppression and more amenable for non polar compounds than ESI but usually lower sensitivity)
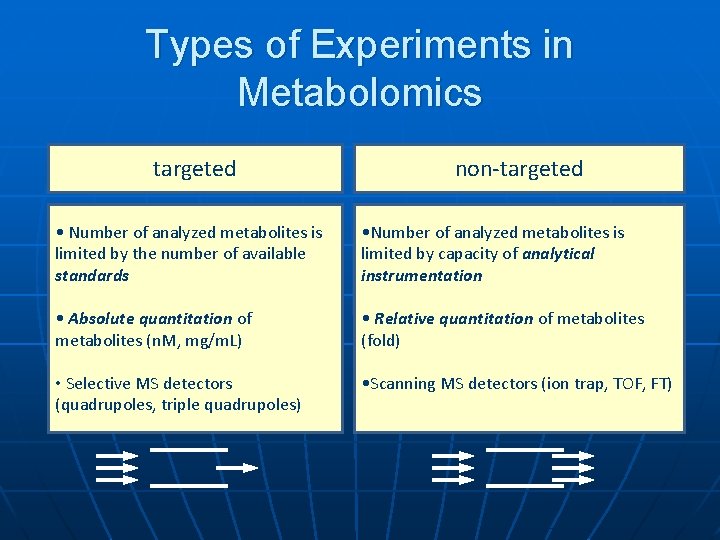
Types of Experiments in Metabolomics targeted non-targeted • Number of analyzed metabolites is limited by the number of available standards • Number of analyzed metabolites is limited by capacity of analytical instrumentation • Absolute quantitation of metabolites (n. M, mg/m. L) • Relative quantitation of metabolites (fold) • Selective MS detectors (quadrupoles, triple quadrupoles) • Scanning MS detectors (ion trap, TOF, FT)
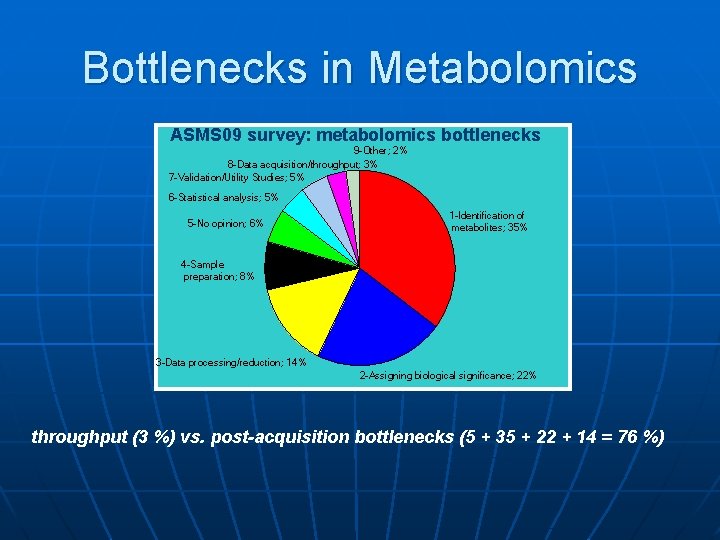
Bottlenecks in Metabolomics ASMS 09 survey: metabolomics bottlenecks 9 -Other; 2% 8 -Data acquisition/throughput; 3% 7 -Validation/Utility Studies; 5% 6 -Statistical analysis; 5% 5 -No opinion; 6% 1 -Identification of metabolites; 35% 4 -Sample preparation; 8% 3 -Data processing/reduction; 14% 2 -Assigning biological significance; 22% throughput (3 %) vs. post-acquisition bottlenecks (5 + 35 + 22 + 14 = 76 %)

GC-MS based metabolomics: overview n 50 - 600 (400) amu mass range mono- and disaccharides, amino acids, fatty acids (mostly primary metabolites) n Derivatization usually required n
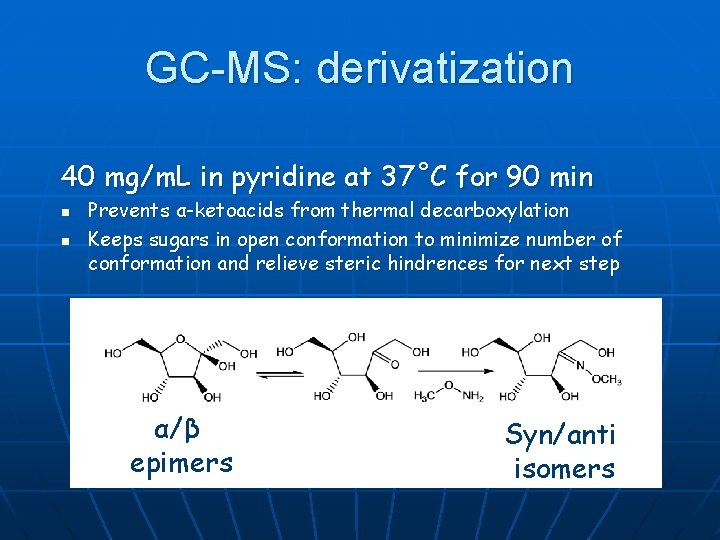
GC-MS: derivatization 40 mg/m. L in pyridine at 37˚C for 90 min n n Prevents α-ketoacids from thermal decarboxylation Keeps sugars in open conformation to minimize number of conformation and relieve steric hindrences for next step α/β epimers Syn/anti isomers
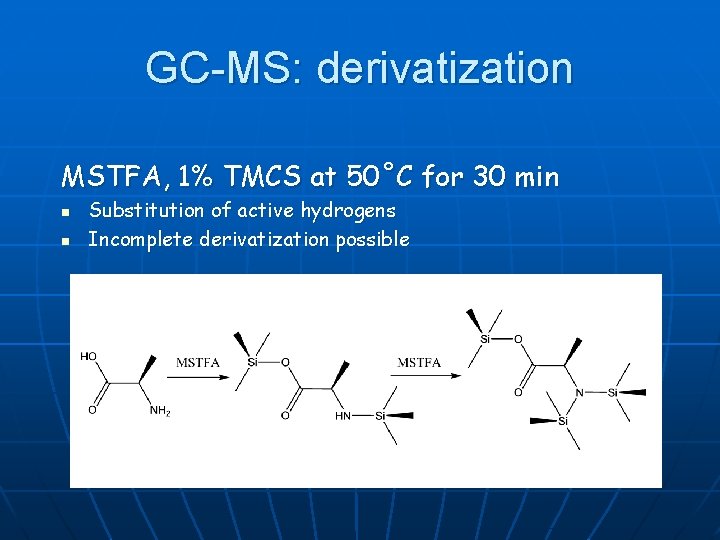
GC-MS: derivatization MSTFA, 1% TMCS at 50˚C for 30 min n n Substitution of active hydrogens Incomplete derivatization possible

GC-MS data analysis
Electron Ionization in GC-MS 70 e. V >> energy of chemical bond n n n Highly reproducible Extensive fragmentation Often no molecular ion observed EI: alpha-cleavage [a ] more common CID MS/MS: inductive cleavage [i ] common
GC-MS: present and future Current GC-MS metabolomics platforms use: 1) nominal resolution mass analyzers (no accurate mass and elemental composition) 2) electron ionization source OE molecular ions, extensive fragmentation, often molecular ion is not observed Advantages: 1) Low cost 2) Good chromatographic separation for many small polar metabolites after derivatization 3) Extensive libraries of fragmentation spectra help identification 4) Retention time is to some extent predictable (retention indices) Trends: 1) Development of high resolution instruments for GC/MS 2) Development of soft ionization sources similar to LC/MS (EE ions, no fragments)
GC-MS data analysis n n n Deconvolution of mass spectra based on chromatographic profiles (e. g freeware AMDIS) Identification of metabolites based on matching to spectral libraries and retention indices Automated processing routines exist for some GC-MS instrument (Setup. X and Bin. Base)
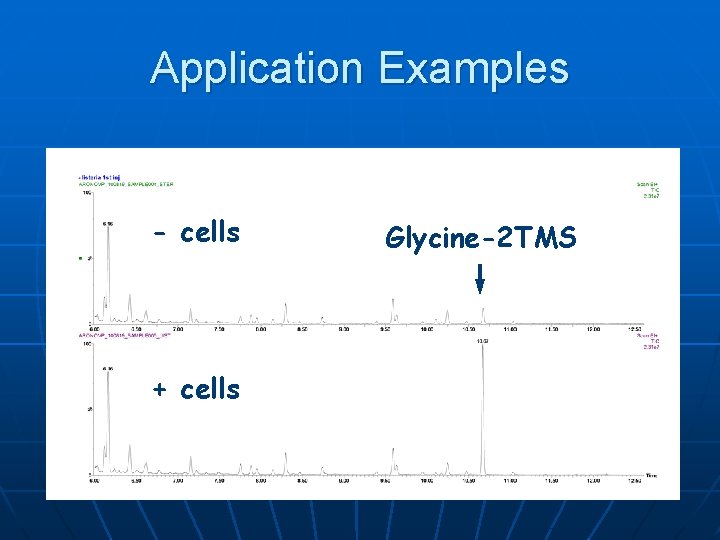
Application Examples - cells + cells Glycine-2 TMS
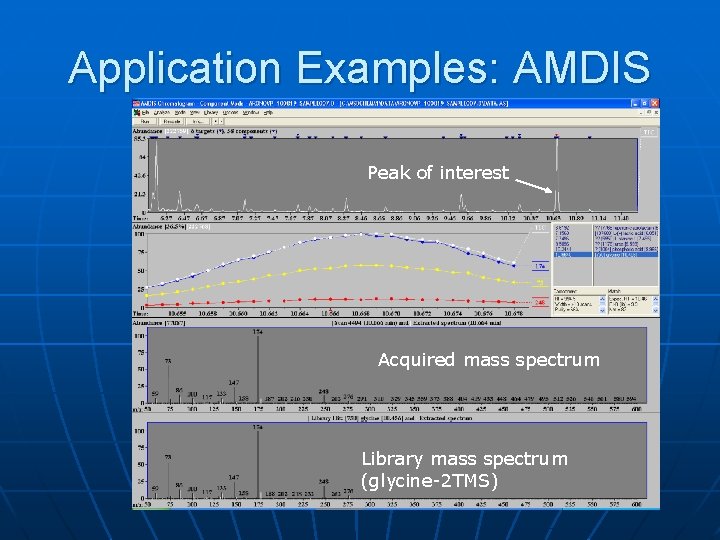
Application Examples: AMDIS Peak of interest Acquired mass spectrum Library mass spectrum (glycine-2 TMS)
LC-MS based metabolomics n n Combination of ionization modes is preferred (ESI, APCI, +, -) Reversed phase LC for non-polar metabolites and hydrophilic interaction chromatography (HILIC) for polar metabolites Detection of spectral “features” (ions) using metabolomics software Identification based on accurate mass, and fragmentation (MS/MS libraries)
![Electrospray Ionization (ESI) R + H+ R– H+ Positive ESI Negative ESI [R+H]+ [R Electrospray Ionization (ESI) R + H+ R– H+ Positive ESI Negative ESI [R+H]+ [R](http://slidetodoc.com/presentation_image/5b981b7dc7264dd81f78087f0bbff489/image-30.jpg)
Electrospray Ionization (ESI) R + H+ R– H+ Positive ESI Negative ESI [R+H]+ [R – H]+
![APCI ESI Soft ionization, pseudomolecular ions [M + H]+, [M - H]- , [M APCI ESI Soft ionization, pseudomolecular ions [M + H]+, [M - H]- , [M](http://slidetodoc.com/presentation_image/5b981b7dc7264dd81f78087f0bbff489/image-31.jpg)
APCI ESI Soft ionization, pseudomolecular ions [M + H]+, [M - H]- , [M + Na]+, [M + Cl]Volatile mobile phase, no inorganic salts (phosphate buffer) Ionization in gas phase Ionization in liquid phase High ionization efficiency for compounds with high proton affinity in gas phase High ionization efficiency for compound ionic in a solution Usually singly charged ions Multiply charged ions common for large biomolecules (proteins, nucleic acids) Compatible with reverse and normal phase, Reverse phase, Mobile phase must be conductive Ion suppression common
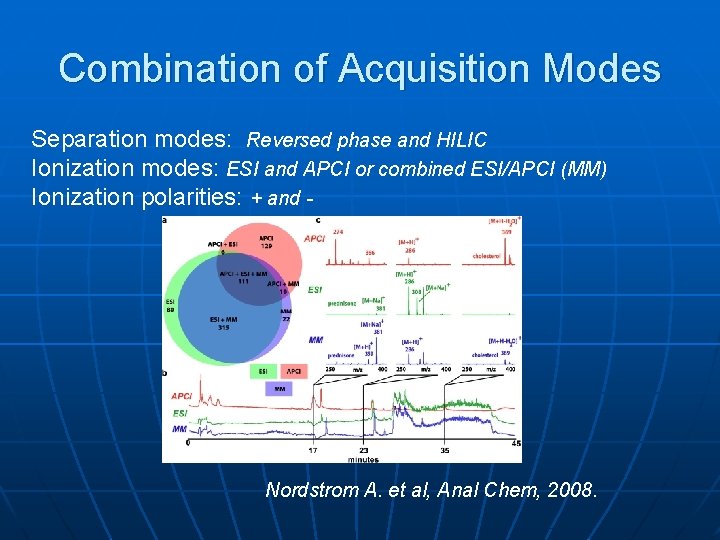
Combination of Acquisition Modes Separation modes: Reversed phase and HILIC Ionization modes: ESI and APCI or combined ESI/APCI (MM) Ionization polarities: + and - Nordstrom A. et al, Anal Chem, 2008.
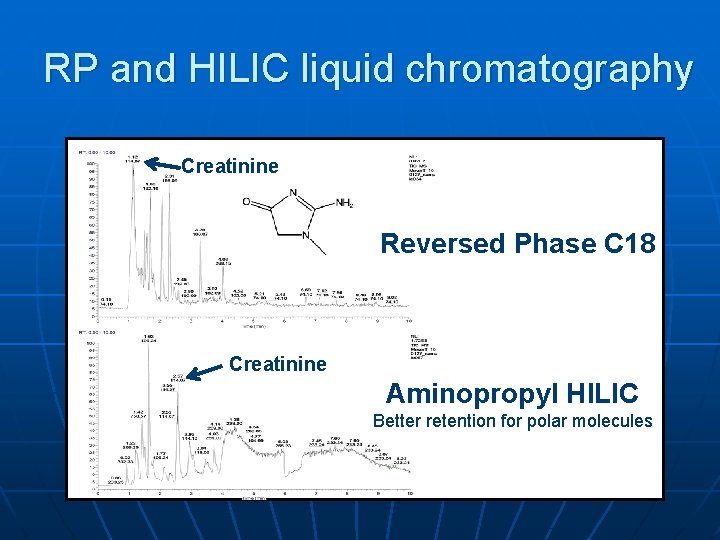
RP and HILIC liquid chromatography Creatinine Reversed Phase C 18 Creatinine Aminopropyl HILIC Better retention for polar molecules
LC-MS: Data Analysis n Alignment of chromatograms (optional) n Detection of ‘features’ in mass chromatograms n n n Removal of isotopic peaks, adducts, fragments etc to improve statistics Statistical analysis Identification based on accurate mass, MS/MS spectra and comparison with standards

Example: Search for bacterial metabolites in humans comparing two groups: controls and people who underwent colectomy (no colon bacteria) Initially software detected 900 features in positive ESI mode After features with missing chromatographic profile were removed 769 features left (visual inspection) After isotopes were removed, 554 features left. Only at this point, these are likely molecular ions of individual metabolites
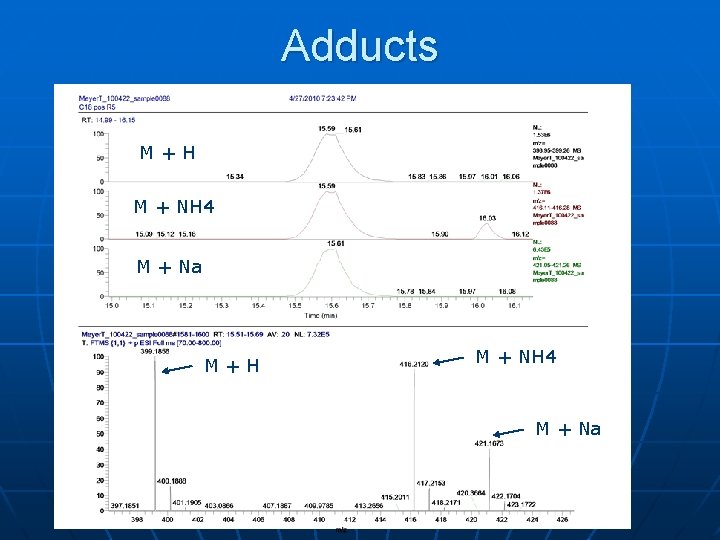
Adducts M+H M + NH 4 M + Na
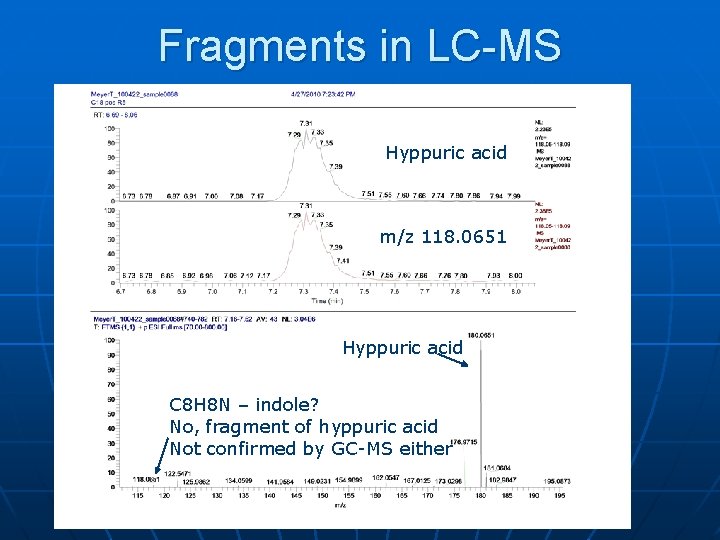
Fragments in LC-MS Hyppuric acid m/z 118. 0651 Hyppuric acid C 8 H 8 N – indole? No, fragment of hyppuric acid Not confirmed by GC-MS either

Identification tools n n Accurate mass search (Bio. Cyc, HMDB, Metlin) MS/MS search (Metlin, Mass. Bank) In addition, many MS manufacturers offer proprietary tools for structure elucidation
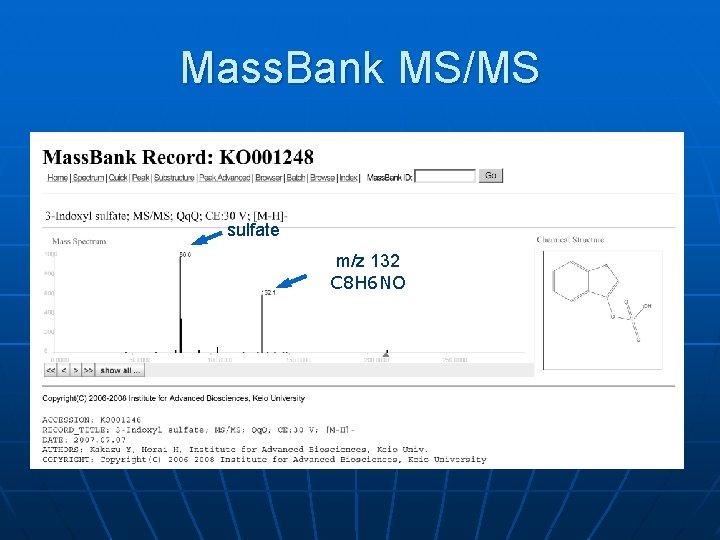
Mass. Bank MS/MS sulfate m/z 132 C 8 H 6 NO
LC-MS Data Analysis Summary n n n Not every peak detected by a mass spectrometer represents an individual metabolite Automated data processing helps to reduce the amount of routine work, however human intervention is still required Accurate mass measurements and MS/MS allow to determine elemental composition of unknowns and their structural components. Confirmation with chemical standards is still required
- Slides: 40