ALUs Functions of ALUs n n Transcriptional Postranscriptional














- Slides: 34

ALUs

Functions of ALUs n n Transcriptional Postranscriptional

Evolution n Emerged 55 million years ago by a fusion of the 5’ and 3’ ends of the 7 SL RNA gene, which encodes the RNA moiety of the signal recognition particle (SRP; SRP and its receptor initiate the transfer of the nascent chain across the endoplasmic reticulum membrane). Primate specific

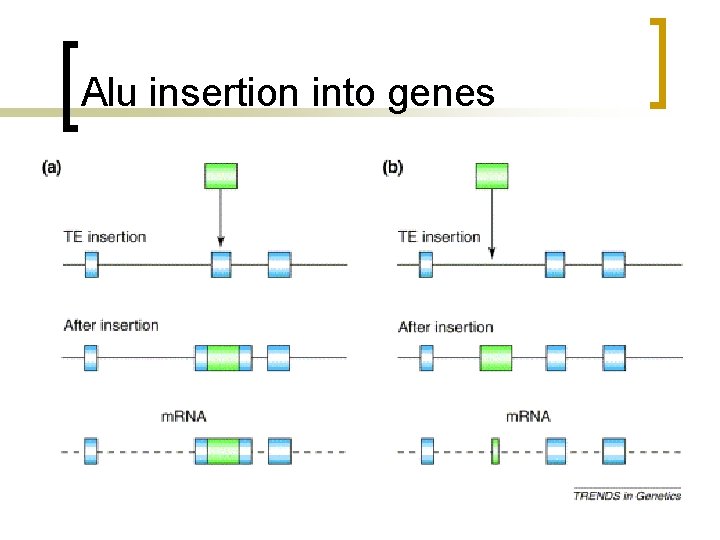
Evolution of genome n n n Insertion into genic regions leads to deleterious mutation Increased rates of mutation, deletions and duplications through non-allelic recombination Provide new regulatory elements into the neighboring genes where they insert (new enhancers, promoters, and poly. A tails)

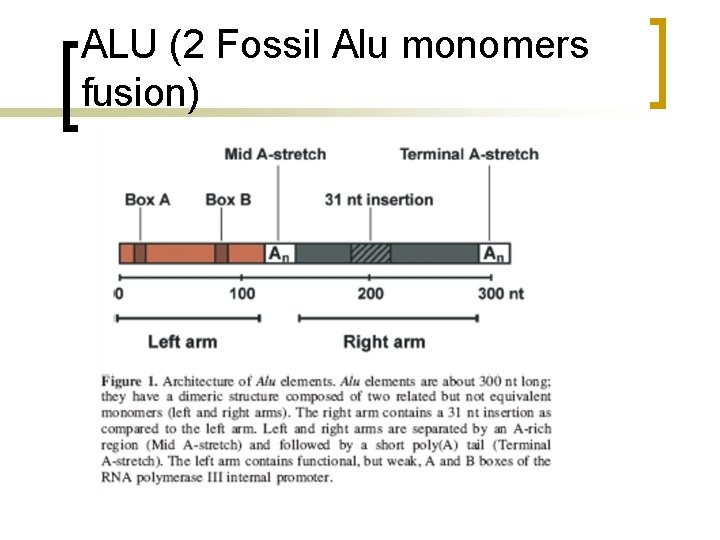
ALU (2 Fossil Alu monomers fusion)

Alu insertion into genes

alternative splicing n A striking example: Drosophila axon guidance receptor gene, Dscam, may potentially generate 38 000 DSCAM isoforms by alternative splicing.

exonization n mutation of pre-existing intronic sequences that result in the recruitment of intronic sequences into coding regions of m. RNAs. This process is called exonization.

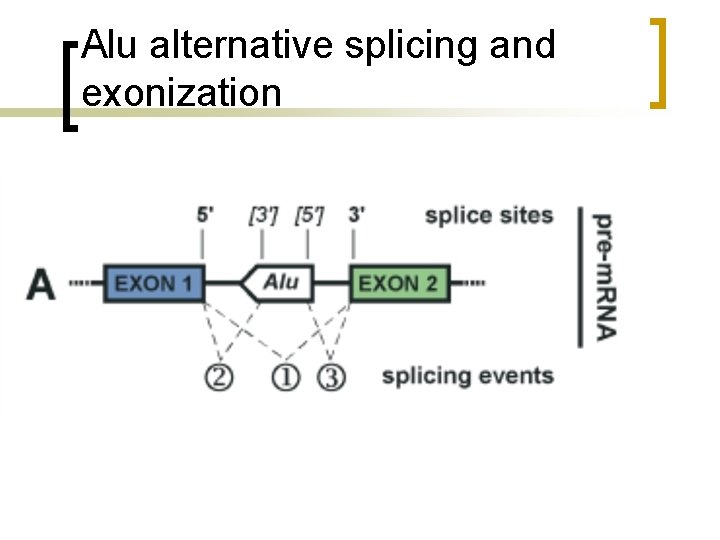
Alu alternative splicing and exonization

Alu n n Alu are present in introns and also in coding regions Alu has several potential splice sites in its sequence

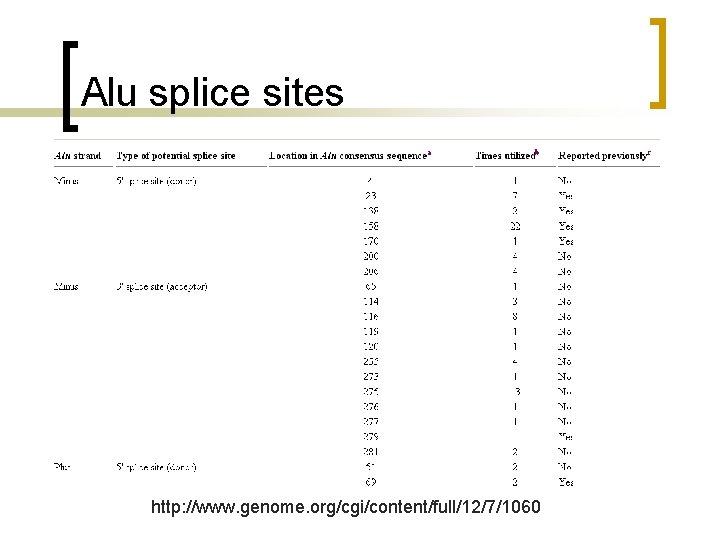
Alu splice sites http: //www. genome. org/cgi/content/full/12/7/1060

Alu splice sites n the most favored sites in the antisense Alu consensus were positions 275 and 279 used as 30 splice site (referred as proximal and distal splice sites, respectively) and position 158 used as 50 splice site.

Potential for disease n insertion of an alternative Alu exon could also lead to a genetic disease. A mutation in the intron 6 of the CTDP 1 gene, which creates an alternatively spliced Alu exon, results in CCFDN (congenital cataracts, facial dysmorphism and neuropathy) syndrome.

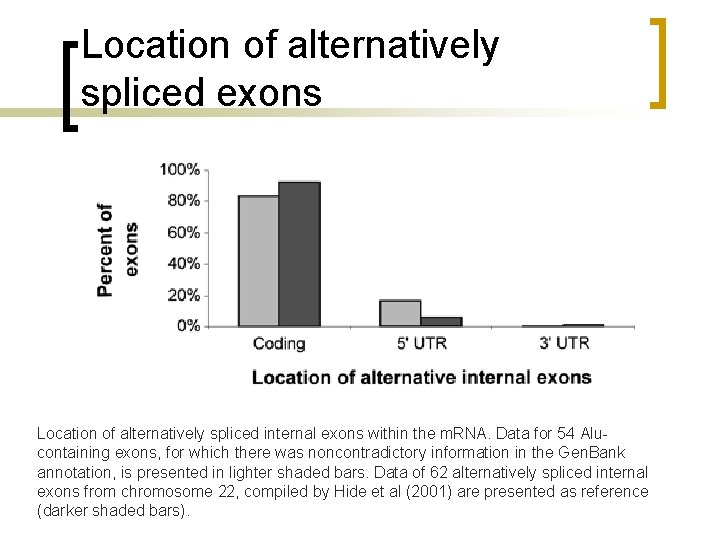
Location of alternatively spliced exons Location of alternatively spliced internal exons within the m. RNA. Data for 54 Alucontaining exons, for which there was noncontradictory information in the Gen. Bank annotation, is presented in lighter shaded bars. Data of 62 alternatively spliced internal exons from chromosome 22, compiled by Hide et al (2001) are presented as reference (darker shaded bars).

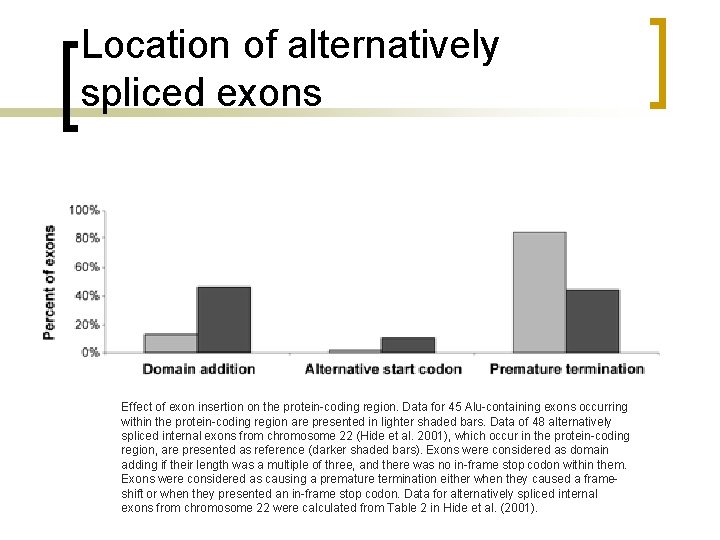
Location of alternatively spliced exons Effect of exon insertion on the protein-coding region. Data for 45 Alu-containing exons occurring within the protein-coding region are presented in lighter shaded bars. Data of 48 alternatively spliced internal exons from chromosome 22 (Hide et al. 2001), which occur in the protein-coding region, are presented as reference (darker shaded bars). Exons were considered as domain adding if their length was a multiple of three, and there was no in-frame stop codon within them. Exons were considered as causing a premature termination either when they caused a frameshift or when they presented an in-frame stop codon. Data for alternatively spliced internal exons from chromosome 22 were calculated from Table 2 in Hide et al. (2001).

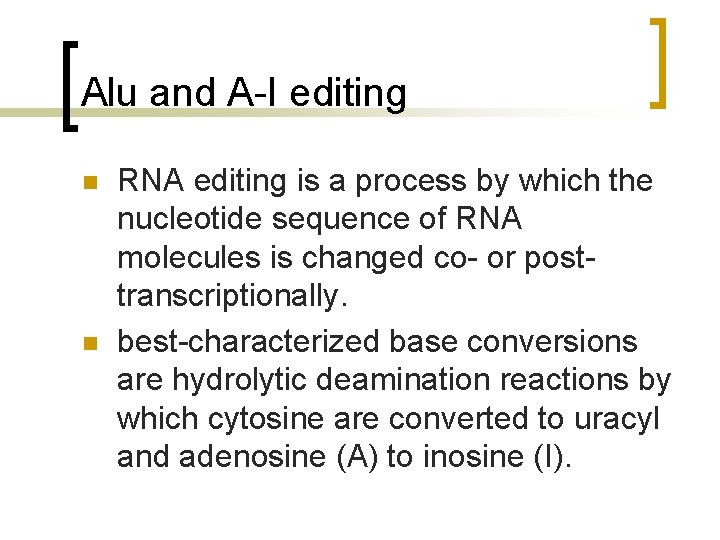
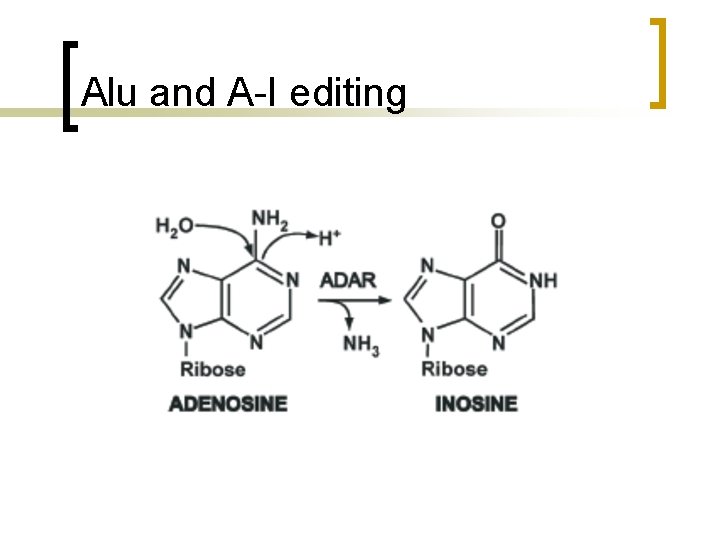
Alu and A-I editing n n RNA editing is a process by which the nucleotide sequence of RNA molecules is changed co- or posttranscriptionally. best-characterized base conversions are hydrolytic deamination reactions by which cytosine are converted to uracyl and adenosine (A) to inosine (I).

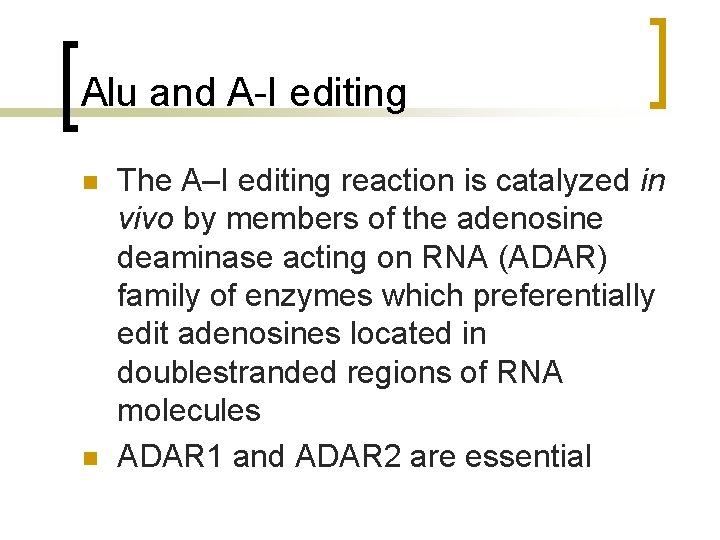
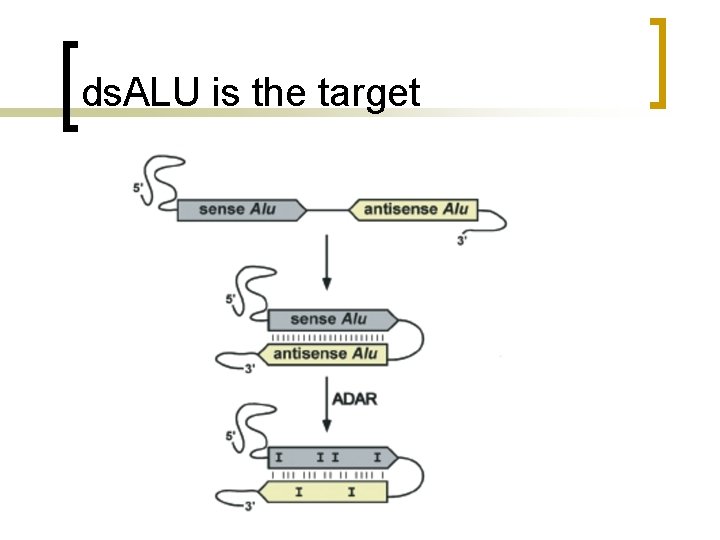
Alu and A-I editing n n The A–I editing reaction is catalyzed in vivo by members of the adenosine deaminase acting on RNA (ADAR) family of enzymes which preferentially edit adenosines located in doublestranded regions of RNA molecules ADAR 1 and ADAR 2 are essential

Alu and A-I editing

Mass of inosine n apparent mass of inosine estimated to one molecule per 17 000 bases in rat brain tissue and one molecule per 33 000 bases in heart tissue

Detecting inosine computationally n aligning m. RNAs (EST and c. DNA databases) to the human genome sequence and detecting A–G substitutions. As I is read as G by sequencing, the presence of A–G substitutions between genomic DNA and m. RNA reflects the presence of an inosine edited site.

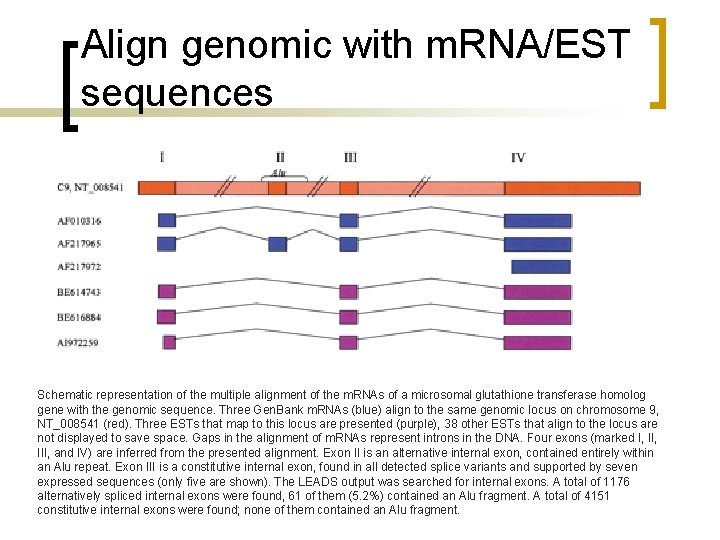
Align genomic with m. RNA/EST sequences Schematic representation of the multiple alignment of the m. RNAs of a microsomal glutathione transferase homolog gene with the genomic sequence. Three Gen. Bank m. RNAs (blue) align to the same genomic locus on chromosome 9, NT_008541 (red). Three ESTs that map to this locus are presented (purple), 38 other ESTs that align to the locus are not displayed to save space. Gaps in the alignment of m. RNAs represent introns in the DNA. Four exons (marked I, III, and IV) are inferred from the presented alignment. Exon II is an alternative internal exon, contained entirely within an Alu repeat. Exon III is a constitutive internal exon, found in all detected splice variants and supported by seven expressed sequences (only five are shown). The LEADS output was searched for internal exons. A total of 1176 alternatively spliced internal exons were found, 61 of them (5. 2%) contained an Alu fragment. A total of 4151 constitutive internal exons were found; none of them contained an Alu fragment.

Findings n n Kim et al. (31) identified 30 085 substitutions in 2674 different transcripts, Levanon et al. (32) identified 12 723 substitutions in 1637 different transcripts, and Athanasiadis et al. (33) found 14 500 substitutions in 1445 m. RNAs. >90% of all A–I substitutions occur within Alu elements contained in m. RNAs.

Findings n editing is favored when a distance <2 kb separates two Alu elements in opposite orientations. These data defined a model in which two closely inserted Alu elements base pair and become an ideal substrate for ADAR

ds. ALU is the target

Alu elements and protein translation n Alu elements contain the internal A and B boxes of the RNA polymerase III promoter from the 7 SL RNA gene. These internal promoter elements significantly diverge from the consensus and are too weak to drive efficient transcription of Alu elements, which is then dependent on sequences flanking their site of insertion.

ALU (2 Fossil Alu monomers fusion)

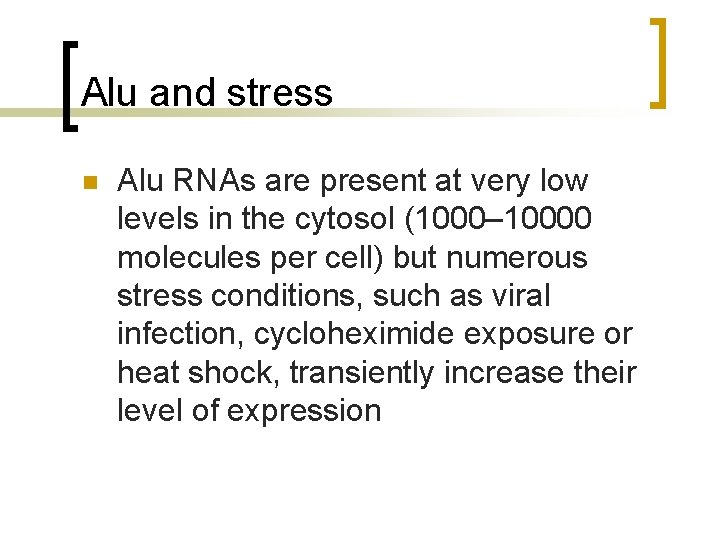
Alu and stress n Alu RNAs are present at very low levels in the cytosol (1000– 10000 molecules per cell) but numerous stress conditions, such as viral infection, cycloheximide exposure or heat shock, transiently increase their level of expression

Alu as translational inhibitors n n They were proposed to bind PKR (double-stranded RNAdependent protein kinase) and regulation translational initiation. But later found that inhibition is dose dependent and may not involve PKR

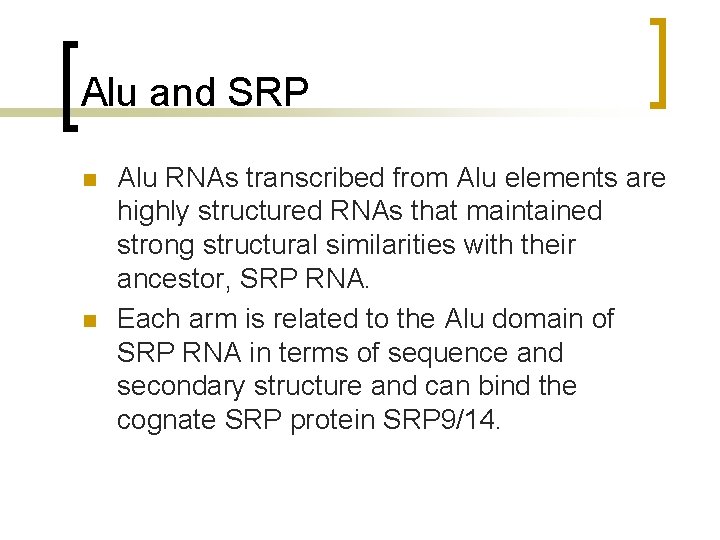
Alu and SRP n n Alu RNAs transcribed from Alu elements are highly structured RNAs that maintained strong structural similarities with their ancestor, SRP RNA. Each arm is related to the Alu domain of SRP RNA in terms of sequence and secondary structure and can bind the cognate SRP protein SRP 9/14.

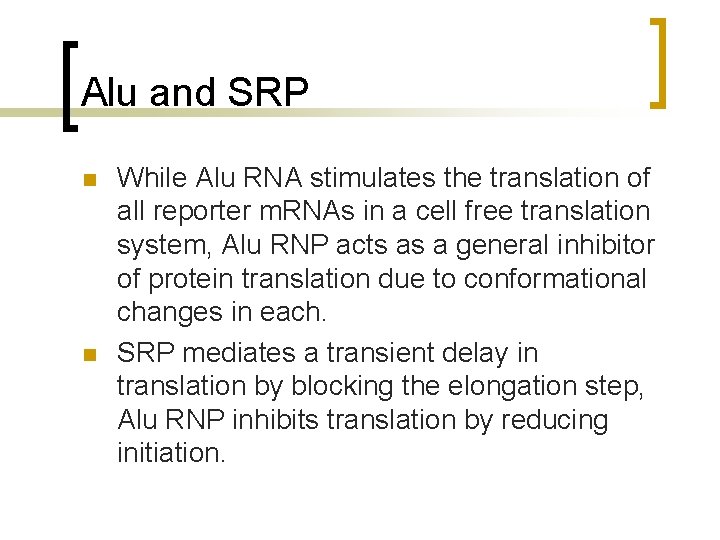
Alu and SRP n n While Alu RNA stimulates the translation of all reporter m. RNAs in a cell free translation system, Alu RNP acts as a general inhibitor of protein translation due to conformational changes in each. SRP mediates a transient delay in translation by blocking the elongation step, Alu RNP inhibits translation by reducing initiation.

BRCA 1 n n Translation regulation by an Alu element in the UTR is BRCA 1 is a DNA repair protein whose mutation is associated with breast cancer. The 80 kb genomic sequence of this gene is composed at 40% of Alu elements.

BRCA 1 n n BRCA 1 m. RNA exists in two forms that differ in their leader sequences and in their patterns of expression. These two transcripts are formed by selective use of different promoters. The isoform with a short 50 -UTR is expressed in normal and cancerous mammary tissue whereas the isoform with a longer 50 -UTR is expressed only in breast cancer tissue.

BRCA 1 n The latter m. RNA is much less efficiently translated than the other one and this translational defect has been shown to be due to an Alu element in the 50 -UTR of this transcript. This Alu element has a 60 nt deletion in the left arm but the right one is intact and forms the stable secondary structure that partially prevents translation initiation.

BRCA 1 n Deregulation of BRCA 1 transcription in cancer then results in a higher proportion of translationally inhibited m. RNA, which contributes to a decrease in the BRCA 1 protein level leading to accumulation of defects and mutations, and ultimately to cancer.
 Basa kang digunakake murid marang guru yaiku basa
Basa kang digunakake murid marang guru yaiku basa Rombi diagonaalid
Rombi diagonaalid Kalimat nyuwun kawigaten
Kalimat nyuwun kawigaten Logaritmi definitsioon
Logaritmi definitsioon Kampus wong alus
Kampus wong alus Alus ja öeldis 8 klass
Alus ja öeldis 8 klass Evaluating functions and operations on functions
Evaluating functions and operations on functions Absolute value functions as piecewise functions
Absolute value functions as piecewise functions Evaluating functions
Evaluating functions Identity parent function
Identity parent function Nature of hrm
Nature of hrm Identities trig
Identities trig Noun functions
Noun functions Seven functions of marketing
Seven functions of marketing Functions of mythology
Functions of mythology Session layer protocols
Session layer protocols Polynomial threshold functions
Polynomial threshold functions Fourier transform of product of two functions
Fourier transform of product of two functions Shape functions
Shape functions Elevated ridges “winding” around the brain
Elevated ridges “winding” around the brain Nose pns
Nose pns Gcse transformations of graphs
Gcse transformations of graphs How do you know if a graph opens up or down
How do you know if a graph opens up or down Define personal selling
Define personal selling Functions of stage lighting
Functions of stage lighting Polynomial
Polynomial Exponential function
Exponential function Integral of piecewise function
Integral of piecewise function Function of plasma membrane
Function of plasma membrane Properties of quadratic function
Properties of quadratic function Cstring member functions
Cstring member functions Kinds of foreign exchange market
Kinds of foreign exchange market Smile verbs
Smile verbs Degree of denominator
Degree of denominator Brics functions
Brics functions