Tools and techniques to manipulate DNA and its


















- Slides: 76

Tools and techniques to manipulate DNA and its products.

Tools for manipulating DNA 1. CUTTING Cutting DNA into fragments using restriction enzymes: these enzymes cut at specific DNA sequences and are only found in prokaryotic organisms 2. PASTING Pasting DNA fragments together using enzymes called DNA ligases. We can join fragments of DNA to make what is called “recombinant DNA”. DNA ligases are found in many species including humans. 3. COPYING Making many copies of DNA (amplification) using heat resistant DNA polymerases in the Polymerase Chain Reaction (PCR) technique. 4. TRANSFERRING DNA is placed into cells using vectors such as “plasmids”. This technique is called transformation in prokaryotic cells.

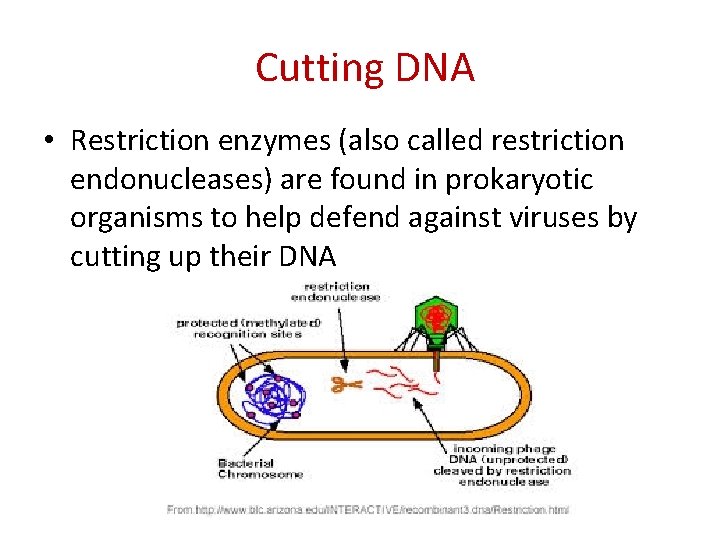
Cutting DNA • Restriction enzymes (also called restriction endonucleases) are found in prokaryotic organisms to help defend against viruses by cutting up their DNA

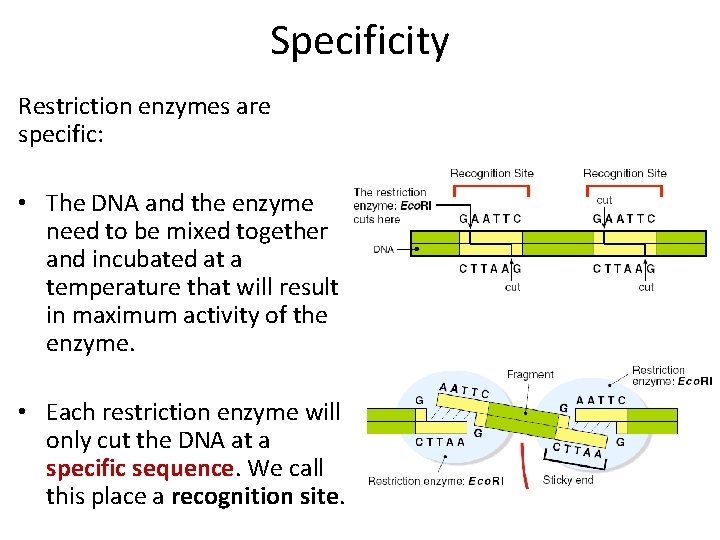
Specificity Restriction enzymes are specific: • The DNA and the enzyme need to be mixed together and incubated at a temperature that will result in maximum activity of the enzyme. • Each restriction enzyme will only cut the DNA at a specific sequence. We call this place a recognition site.

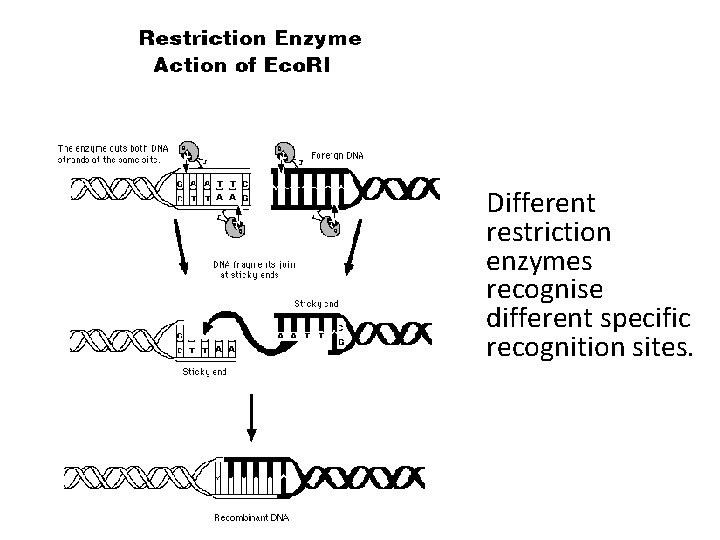
Different restriction enzymes recognise different specific recognition sites.

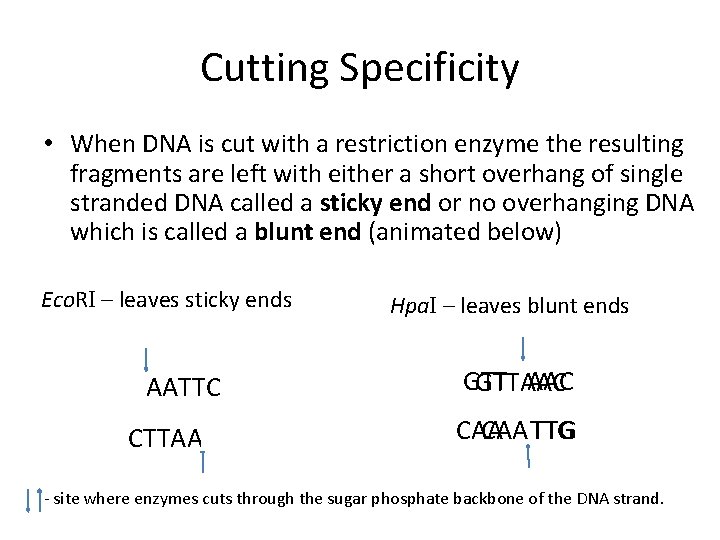
Cutting Specificity • When DNA is cut with a restriction enzyme the resulting fragments are left with either a short overhang of single stranded DNA called a sticky end or no overhanging DNA which is called a blunt end (animated below) Eco. RI – leaves sticky ends Hpa. I – leaves blunt ends GAATTC GTT AAC GTTAAC CTTAAG CAA TTG - site where enzymes cuts through the sugar phosphate backbone of the DNA strand.

Restriction Enzyme Animations • Restriction enzyme (restriction endonuclease) animation: • http: //highered. mcgrawhill. com/olcweb/cgi/pluginpop. cgi? it=swf: : 535: : /sites/dl/ free/0072437316/120078/bio 37. swf: : Restriction%20 Endonuc leases • Restriction enzyme tutorial: • http: //www. dnalc. org/resources/animations/restriction. html

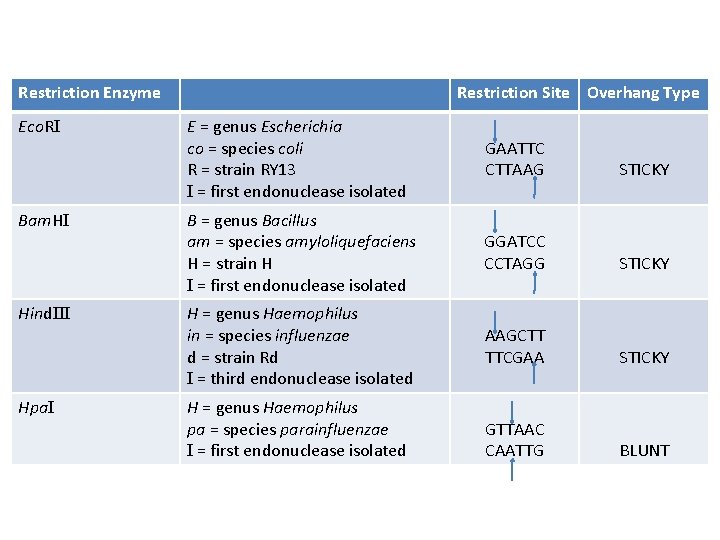
What’s in a name! • Restriction enzymes are named after the organism from which they were isolated. – E. g. Escherichia coli Eco. RI • The Roman number indicates the order of discovery • If another letter is placed in front of the Roman number it signifies a particular strain of the bacterium. R = resistance

Restriction Enzyme Eco. RI Bam. HI Hind. III Hpa. I Restriction Site Overhang Type E = genus Escherichia co = species coli R = strain RY 13 I = first endonuclease isolated GAATTC CTTAAG STICKY B = genus Bacillus am = species amyloliquefaciens H = strain H I = first endonuclease isolated GGATCC CCTAGG STICKY H = genus Haemophilus in = species influenzae d = strain Rd I = third endonuclease isolated AAGCTT TTCGAA STICKY GTTAAC CAATTG BLUNT H = genus Haemophilus pa = species parainfluenzae I = first endonuclease isolated

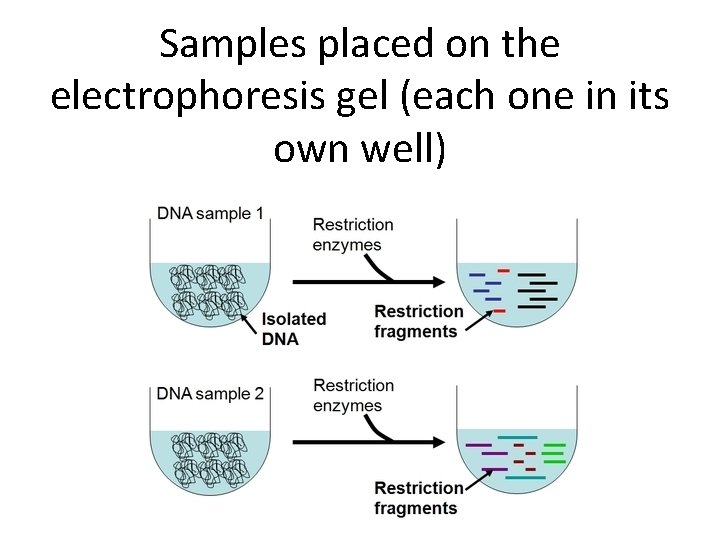
DNA Fragments are sorted according to size by Gel Electrophoresis • The DNA of an organism can be cut up using a mixture of different restriction enzymes • These DNA fragments are then separated into bands containing fragments of the same length (i. e. they have the same number of base pairs - complementary bases joined together in a row)

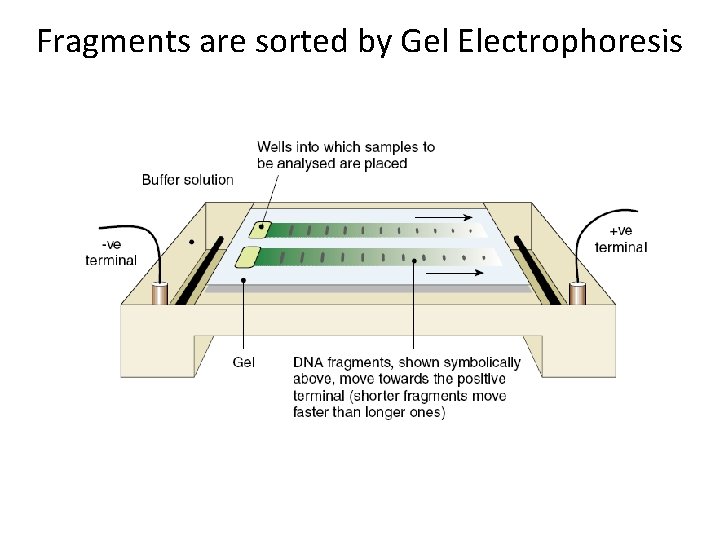
Gel Electrophoresis • The DNA fragments are run through an agarose gel. A current is put through the gel. • DNA is negatively charged and will be drawn towards the positive terminal. • Smaller fragments will move faster while larger fragment will take more time to move through the gel

Samples placed on the electrophoresis gel (each one in its own well)

Fragments are sorted by Gel Electrophoresis

Gel Electrophoresis Animations • https: //www. dnalc. org/resources/animations/ gelelectrophoresis. html • http: //learn. genetics. utah. edu/content/labs/g el/

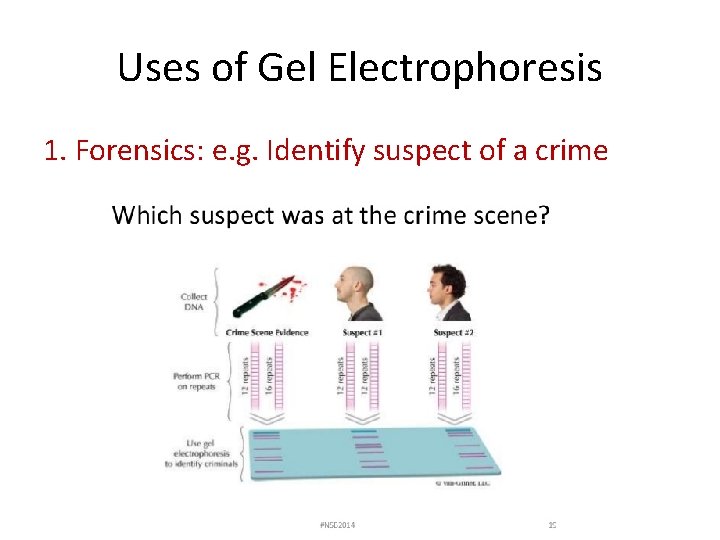
Uses of Gel Electrophoresis 1. Forensics: e. g. Identify suspect of a crime


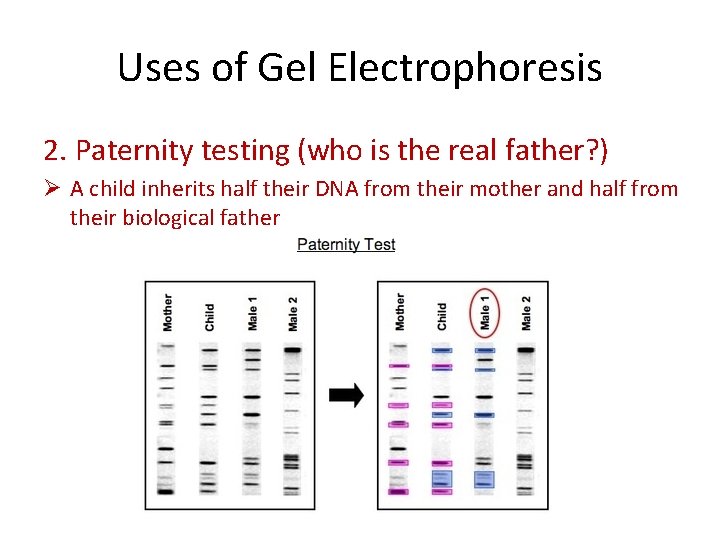
Uses of Gel Electrophoresis 2. Paternity testing (who is the real father? ) Ø A child inherits half their DNA from their mother and half from their biological father

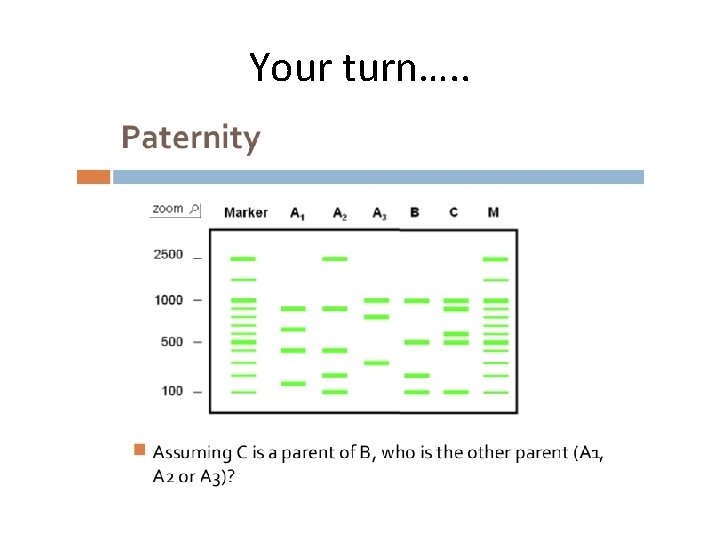
Your turn…. .

Answer • A 2 as B shares DNA with C and with the other parent. The only band that B does not share with C is their 3 rd band, so this must come from the other parent. A 2 is the only one with this matching band.

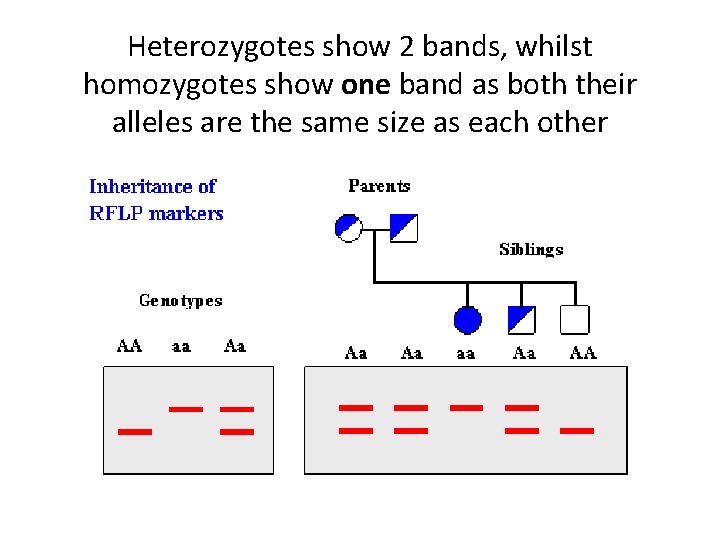
Uses of Gel Electrophoresis 3. Genotyping: • Identification of carriers of recessive or abnormal alleles (which may contribute to genetic disorders) Ø The alleles must be a different size for gel electrophoresis to separate them

Heterozygotes show 2 bands, whilst homozygotes show one band as both their alleles are the same size as each other

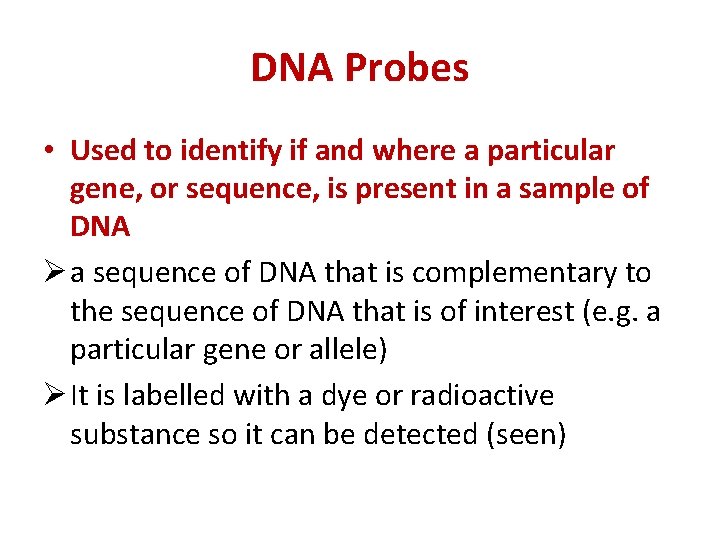
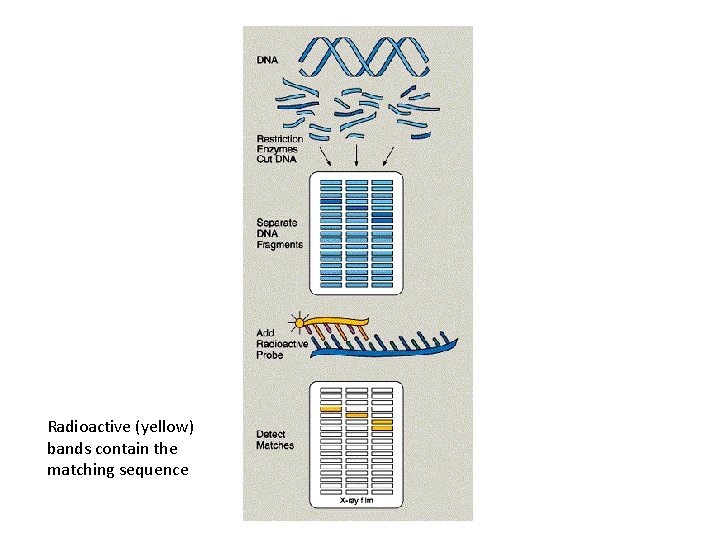
DNA Probes • Used to identify if and where a particular gene, or sequence, is present in a sample of DNA Ø a sequence of DNA that is complementary to the sequence of DNA that is of interest (e. g. a particular gene or allele) Ø It is labelled with a dye or radioactive substance so it can be detected (seen)

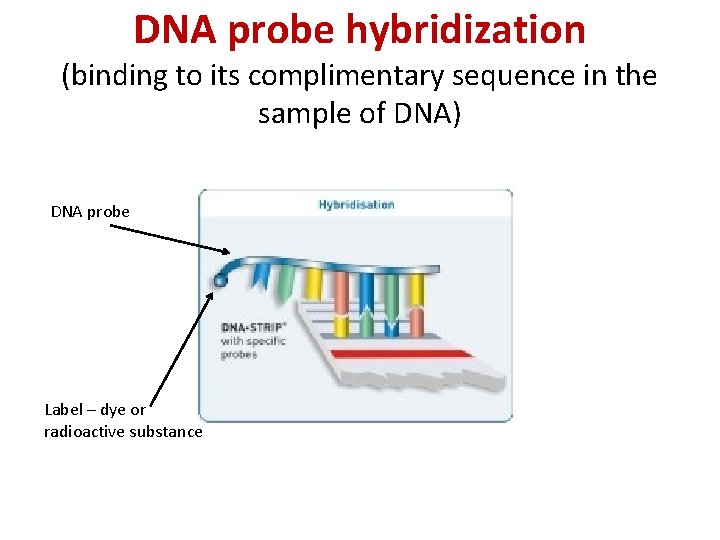
DNA probe hybridization (binding to its complimentary sequence in the sample of DNA) DNA probe Label – dye or radioactive substance

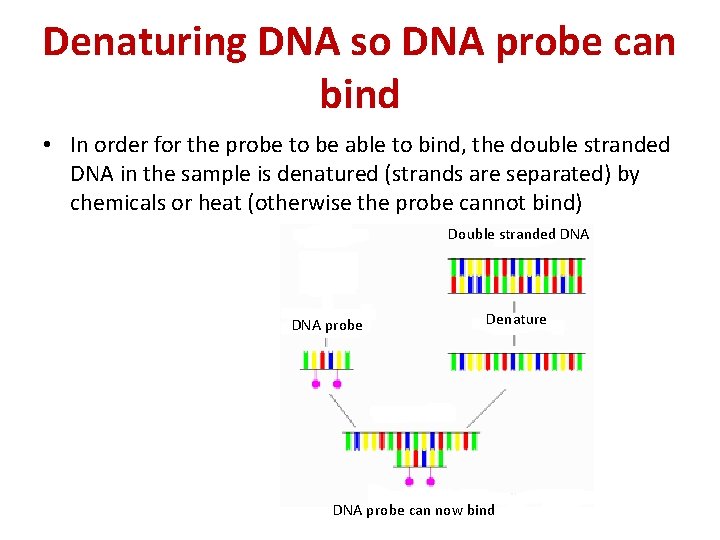
Denaturing DNA so DNA probe can bind • In order for the probe to be able to bind, the double stranded DNA in the sample is denatured (strands are separated) by chemicals or heat (otherwise the probe cannot bind) Double stranded DNA probe Denature DNA probe can now bind

Radioactive (yellow) bands contain the matching sequence

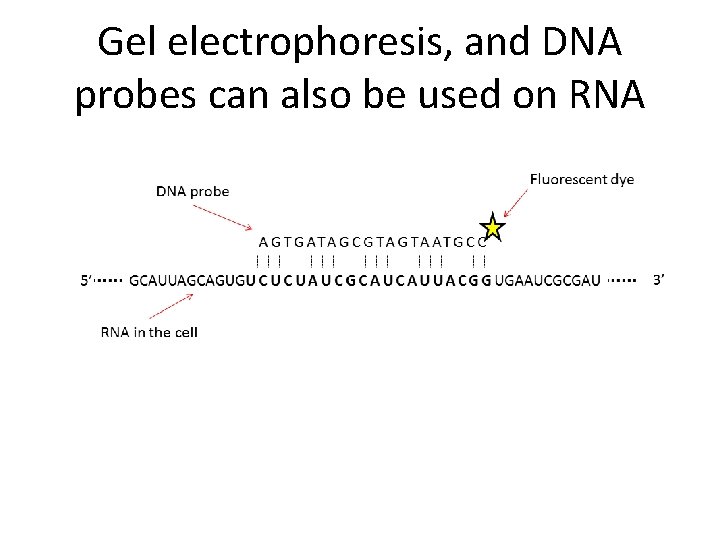
Gel electrophoresis, and DNA probes can also be used on RNA

Gel Electrophoresis and DNA Probe Animations This animation begins with an explanation of gel electrophoresis then ends with DNA probes: • http: //www. sumanasinc. com/webcontent/ani mations/content/gelelectrophoresis. html DNA probes: • http: //sites. sinauer. com/cooper 6 e/animation 0410. html

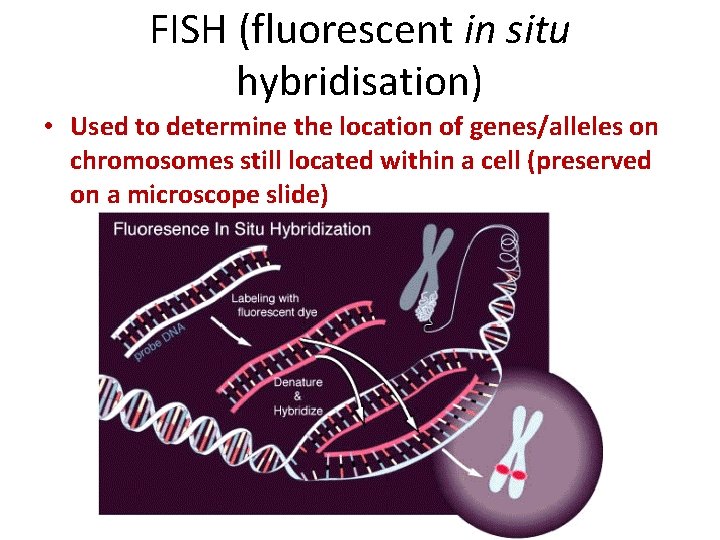
FISH (fluorescent in situ hybridisation) • Used to determine the location of genes/alleles on chromosomes still located within a cell (preserved on a microscope slide)

FISH (fluorescent in situ hybridisation) photos

Copying (Amplification) of DNA using the Polymerase Chain Reaction • Often the sample of DNA obtained from a person, cell or from a crime scene, is too little to work with • The DNA collected is amplified (copied multiple times) using a process called the Polymerase Chain Reaction (PCR)

Uses of PCR • To amplify a small amount of DNA into an analysable quantity – E. g. crime scene, fossils, cancer cells (for research) etc

PCR Ingredients • A heat tolerant DNA Polymerase: Taq DNA Polymerase – is an enzyme that works well at 72°C. • Free nucleotides (single, individual nucleotides used to make the copies of the DNA sequence of interest) • DNA sequence to be copied

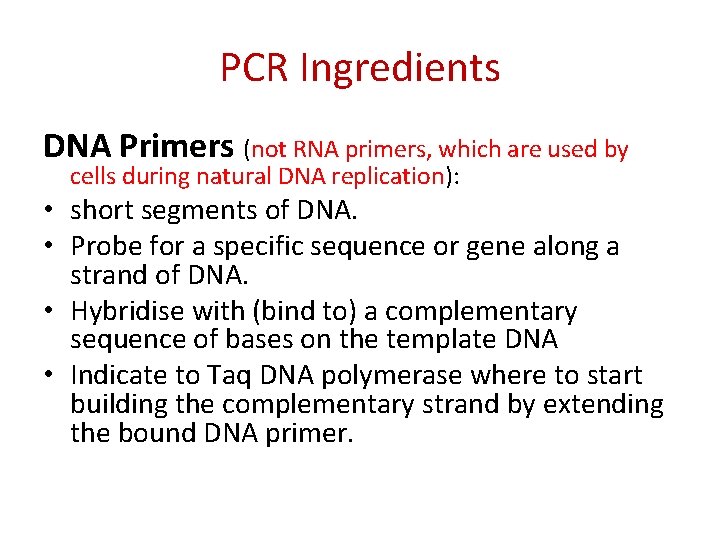
PCR Ingredients DNA Primers (not RNA primers, which are used by cells during natural DNA replication): • short segments of DNA. • Probe for a specific sequence or gene along a strand of DNA. • Hybridise with (bind to) a complementary sequence of bases on the template DNA • Indicate to Taq DNA polymerase where to start building the complementary strand by extending the bound DNA primer.

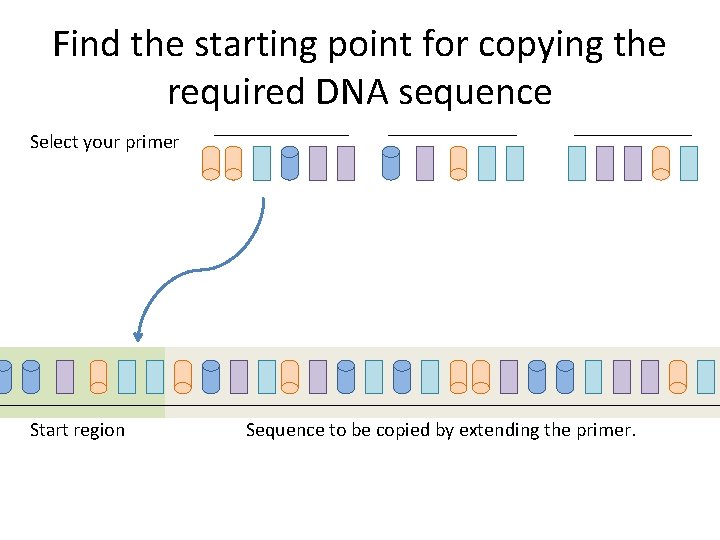
Find the starting point for copying the required DNA sequence Select your primer Start region Sequence to be copied by extending the primer.

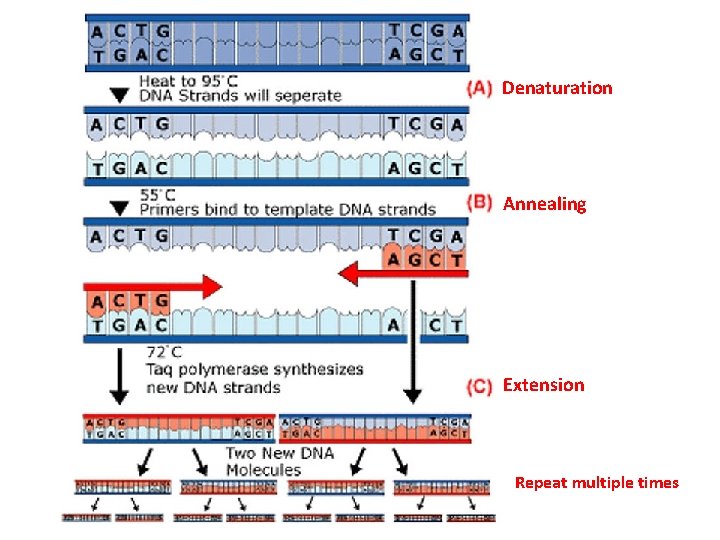
Denaturation Annealing Extension Repeat multiple times

PCR Animations PCR animation: • http: //www. sumanasinc. com/webcontent/ani mations/content/pcr. html A slightly simpler one with a quiz: • http: //highered. mcgrawhill. com/sites/0072556781/student_view 0/ch apter 14/animation_quiz_6. html

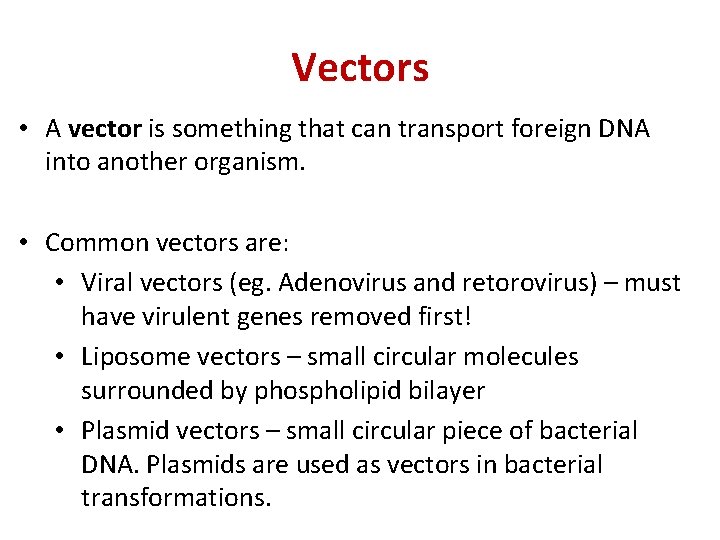
Vectors • A vector is something that can transport foreign DNA into another organism. • Common vectors are: • Viral vectors (eg. Adenovirus and retorovirus) – must have virulent genes removed first! • Liposome vectors – small circular molecules surrounded by phospholipid bilayer • Plasmid vectors – small circular piece of bacterial DNA. Plasmids are used as vectors in bacterial transformations.

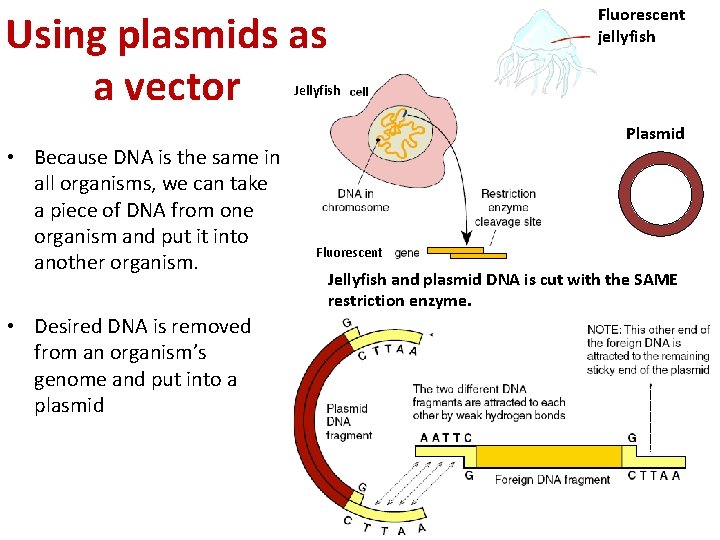
Fluorescent jellyfish Using plasmids as a vector Jellyfish • Because DNA is the same in all organisms, we can take a piece of DNA from one organism and put it into another organism. • Desired DNA is removed from an organism’s genome and put into a plasmid Plasmid Fluorescent Jellyfish and plasmid DNA is cut with the SAME restriction enzyme.

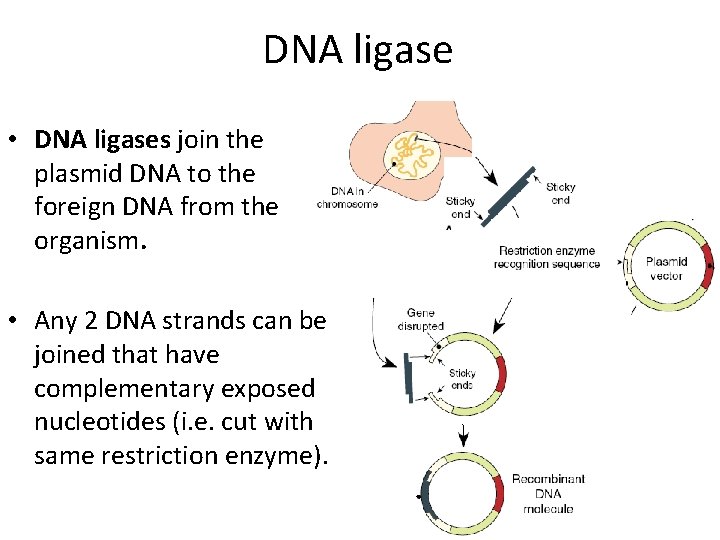
DNA ligase • DNA ligases join the plasmid DNA to the foreign DNA from the organism. • Any 2 DNA strands can be joined that have complementary exposed nucleotides (i. e. cut with same restriction enzyme).

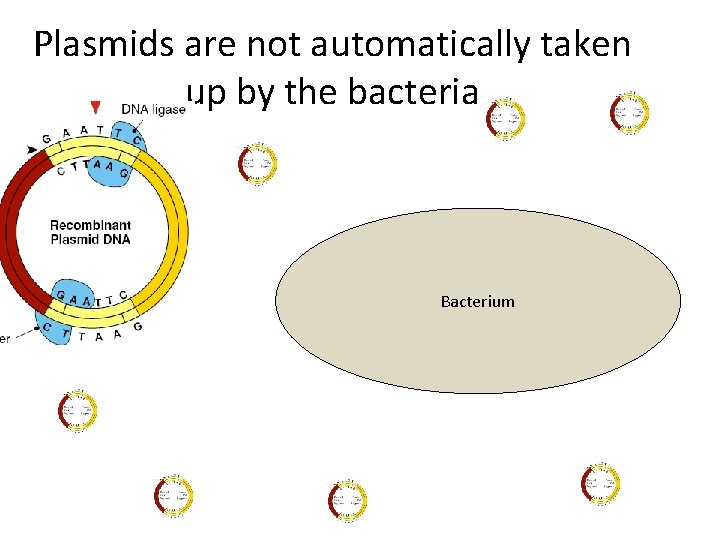
Plasmids are not automatically taken up by the bacteria Bacterium

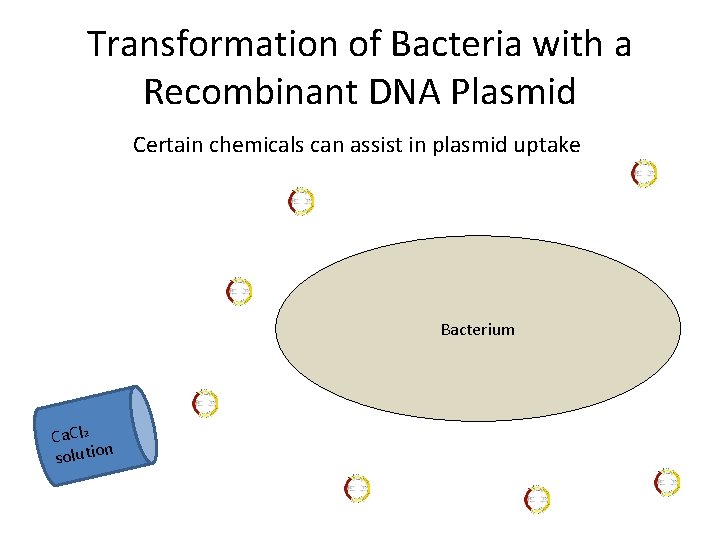
Transformation of Bacteria with a Recombinant DNA Plasmid Certain chemicals can assist in plasmid uptake Bacterium Ca. Cl 2 n solutio

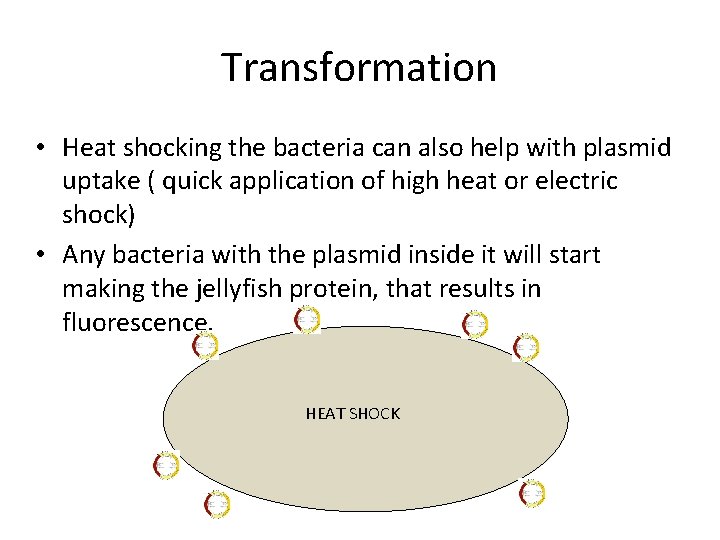
Transformation • Heat shocking the bacteria can also help with plasmid uptake ( quick application of high heat or electric shock) • Any bacteria with the plasmid inside it will start making the jellyfish protein, that results in fluorescence. HEAT SHOCK

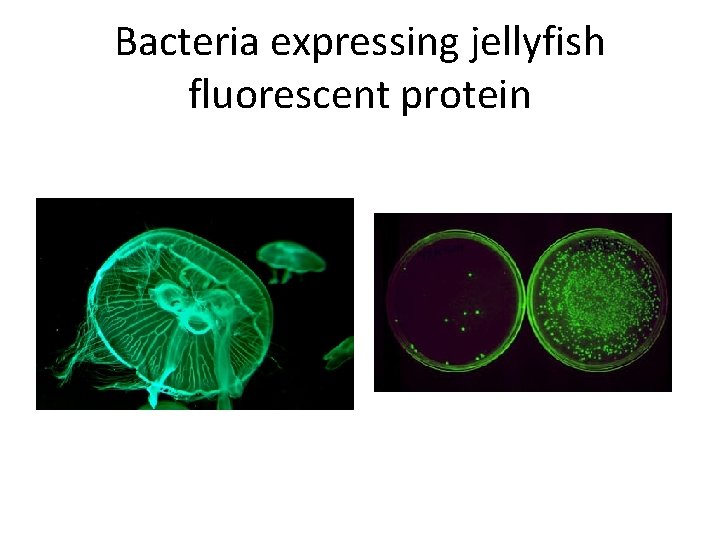
Bacteria expressing jellyfish fluorescent protein

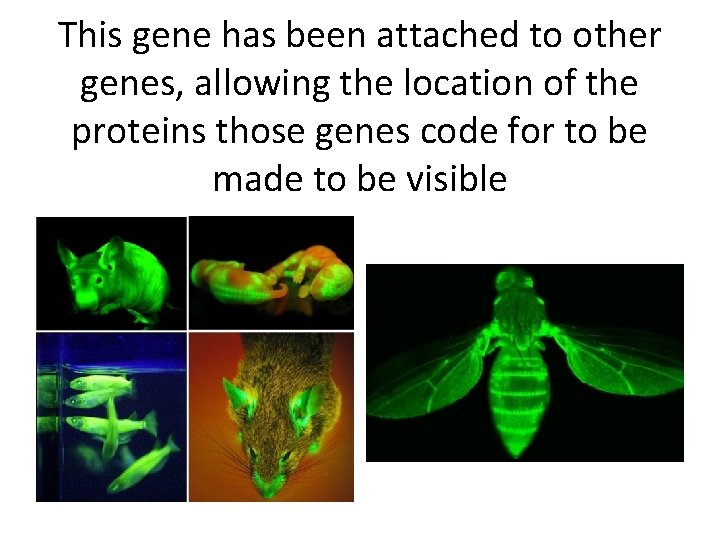
This gene has been attached to other genes, allowing the location of the proteins those genes code for to be made to be visible

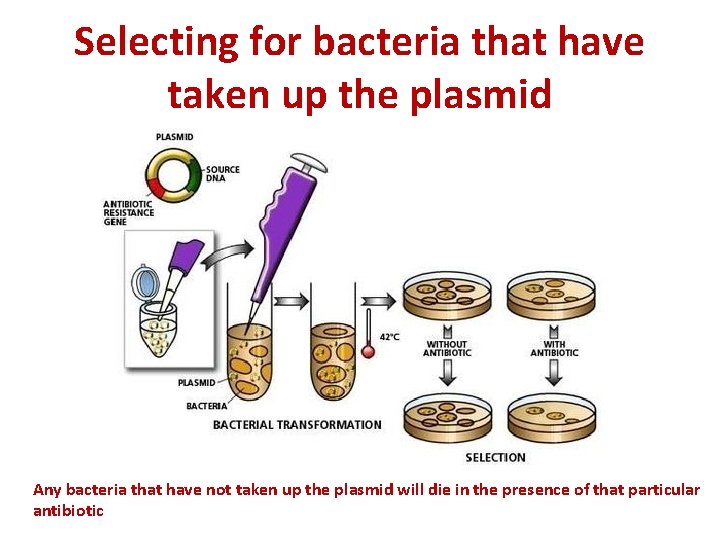
Selecting for bacteria that have taken up the plasmid • Not all bacteria take up the plasmid • To determine which bacteria have taken up the plasmid, selection markers are used (DNA which codes for resistance to a particular antibiotic or resistance to high copper levels, or some other marker such as a fluorescent protein) • The DNA coding for the selection marker is included in the plasmid.

Selecting for bacteria that have taken up the plasmid Any bacteria that have not taken up the plasmid will die in the presence of that particular antibiotic

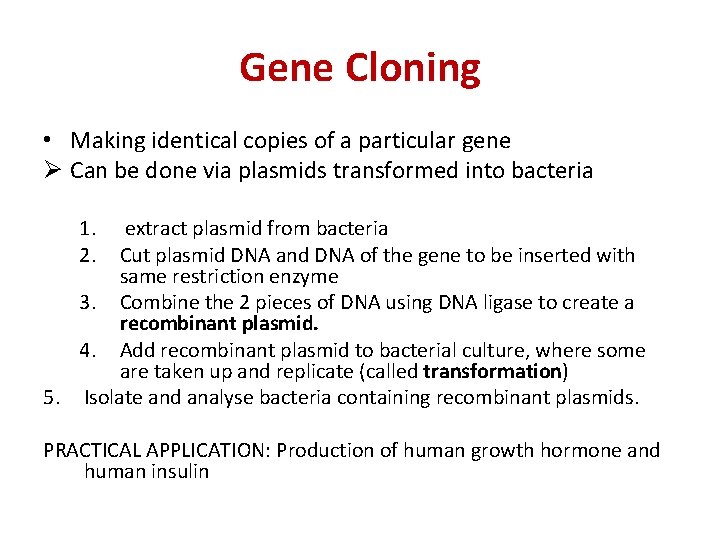
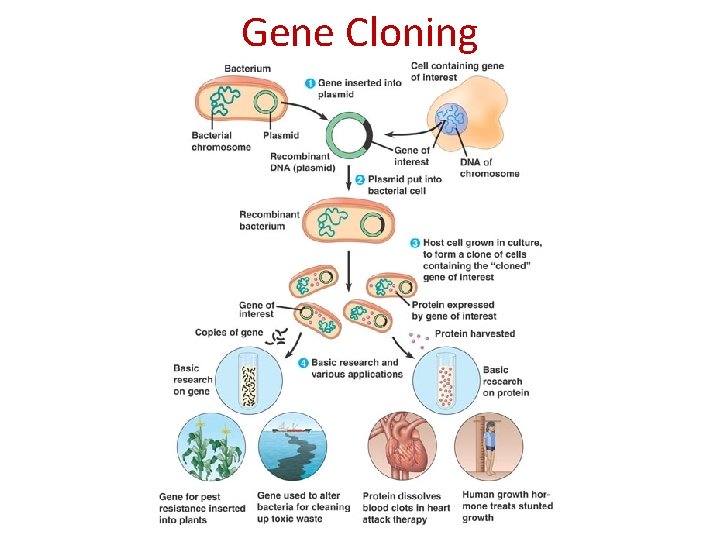
Gene Cloning • Making identical copies of a particular gene Ø Can be done via plasmids transformed into bacteria 1. 2. extract plasmid from bacteria Cut plasmid DNA and DNA of the gene to be inserted with same restriction enzyme 3. Combine the 2 pieces of DNA using DNA ligase to create a recombinant plasmid. 4. Add recombinant plasmid to bacterial culture, where some are taken up and replicate (called transformation) 5. Isolate and analyse bacteria containing recombinant plasmids. PRACTICAL APPLICATION: Production of human growth hormone and human insulin

Gene Cloning

Gene Cloning Animation • http: //highered. mheducation. com/olcweb/cgi /pluginpop. cgi? it=swf: : 535: : /sites/dl/free /0072437316/120078/micro 10. swf: : Steps+in+ Cloning+a+Gene

Transgenic Organisms (TGO’s) • Organisms that contain a gene(s) of a different organism (e. g. tomato plants with frost resistant genes from salmon; cows that produce human breast milk; soy plants containing genes to provide herbicide resistance and plants that produce vaccines against human diseases)

Transgenic Organisms (TGO’s) • Foreign gene can be introduced via either: I. Plasmid with foreign gene placed into organism’s stem/embryonic cells II. Nanoparticles coated with DNA shot into cells III. Gene placed into fertilized egg nucleus or embryonic stem cells which are placed into a female uterus



For fun: 12 Bizzare examples of genetic engineering http: //www. mnn. com/green-tech/research-innovations/photos/12 -bizarre-examples-ofgenetic-engineering/mad-science

Genetically Modified Organisms (GMO’s) • Genes have been altered in some way (e. g. nucleotide base changes) • Some GMO’s are also TGO’s (but not all GMO’s have genes from other organisms) • GMO’s that have an existing gene altered result in a modified protein (not a new protein, unlike TGO’s).

Potential Problems with GMO’s • Creation of new toxins and allergens, and increased risk of development of some cancers (http: //www. ncbi. nlm. nih. gov/pubmed/18989835) • Decreased biodiversity due to monocultures (growth of genetically similar, or identical plants). Less resistance to environmental changes. (http: //www. chgeharvard. org/topic/biodiversity-andagriculture) • https: //www. youtube. com/watch? v=1 H 9 WZGKQe. Yg&feature =related

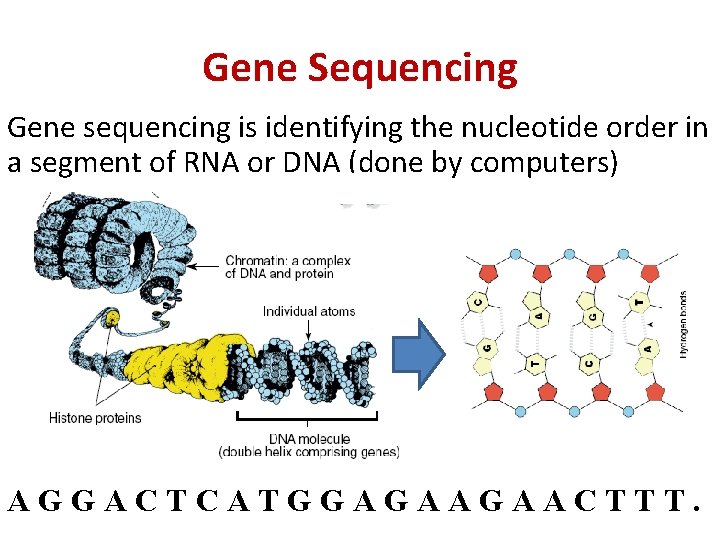
Gene Sequencing Gene sequencing is identifying the nucleotide order in a segment of RNA or DNA (done by computers) AGGACTCATGGAGAAGAACTTT.

DNA Profiling • Compares base sequence of 2 or more individuals • Short tandem repeats (STRs) and variable nucleotide tandem repeats (VNTRs): non-coding sections of DNA repeated many times between genes – E. g. GAGAGAGA • There are more than 10, 000 STR loci in one set of human chromosomes!

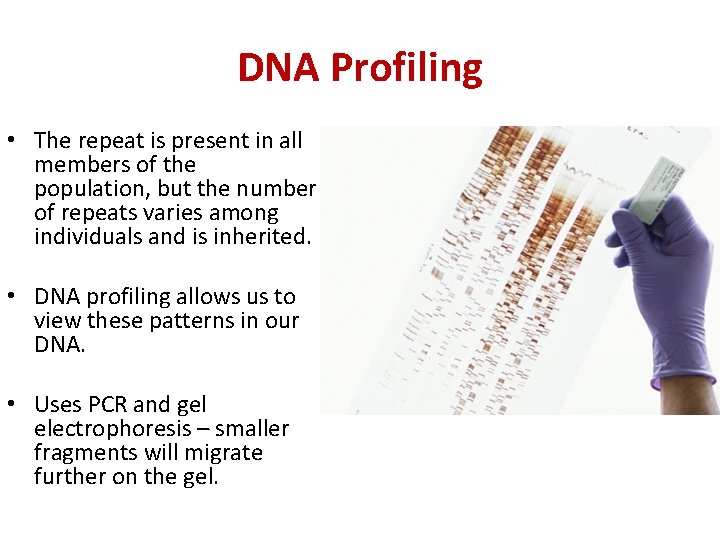
DNA Profiling • The repeat is present in all members of the population, but the number of repeats varies among individuals and is inherited. • DNA profiling allows us to view these patterns in our DNA. • Uses PCR and gel electrophoresis – smaller fragments will migrate further on the gel.

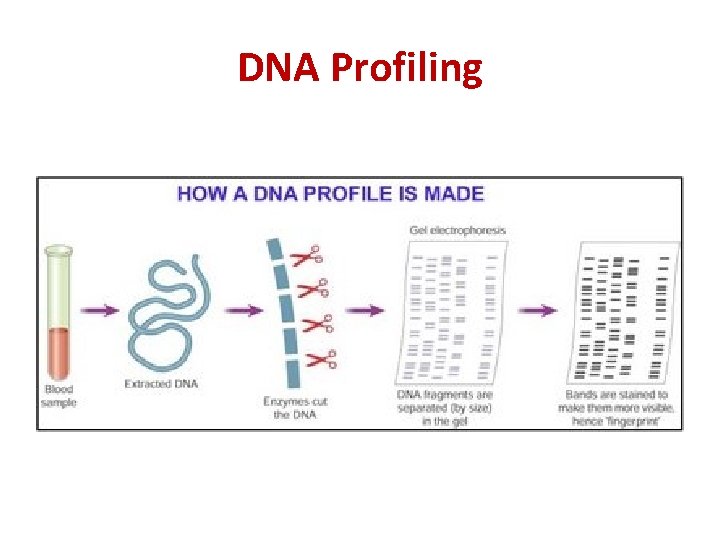
DNA Profiling

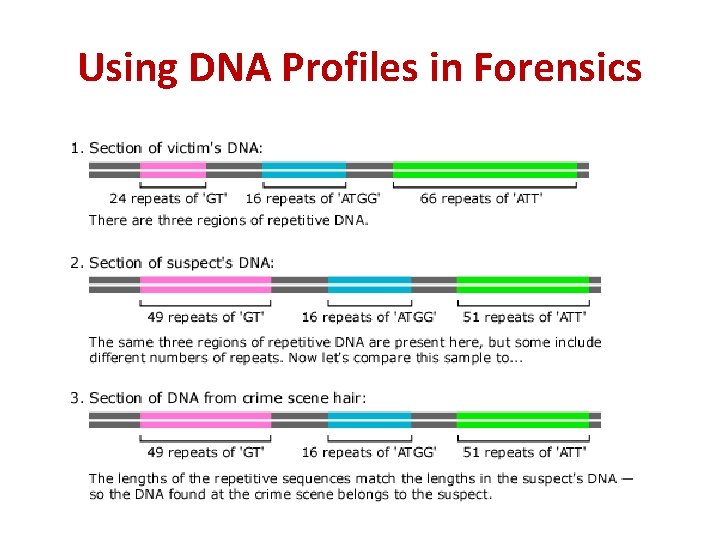
Using DNA Profiles in Forensics

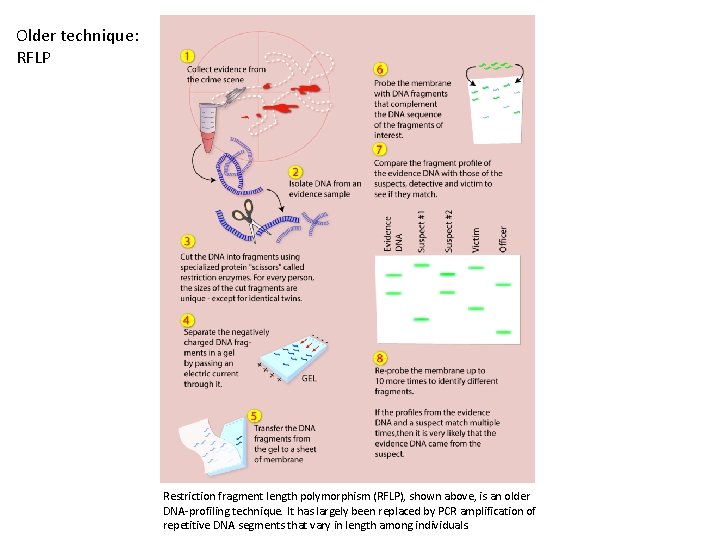
Older technique: RFLP Restriction fragment length polymorphism (RFLP), shown above, is an older DNA-profiling technique. It has largely been replaced by PCR amplification of repetitive DNA segments that vary in length among individuals.


Fun fact – what if an identical twin commits a crime? • Both have identical DNA, but their fingerprints are different (The pattern of our fingerprints is set up by our genes but how those genes express themselves, and therefore the fingerprint pattern they give rise to, is determined by how we touch the amniotic sac between 6 – 13 weeks). • Identical twins will differ in how their fingerprint genes were expressed but they will still have exactly the same fingerprint genes (but different fingerprints) • Current technologies examine differences in identical twin DNA due to individual differences in mutations in each twin’s DNA that occur due to environmental influences and errors in DNA replication

Sounds easy? • On average we accumulate 100 mutations out of 3 billion base pairs in our life (not many overall…so hard to find and detect) • Not all cells have these changes, so they would need to be able to know what tissue the DNA at a crime scene came from • BUT…one day it will be possible (epigenetic markers may be the key…. )

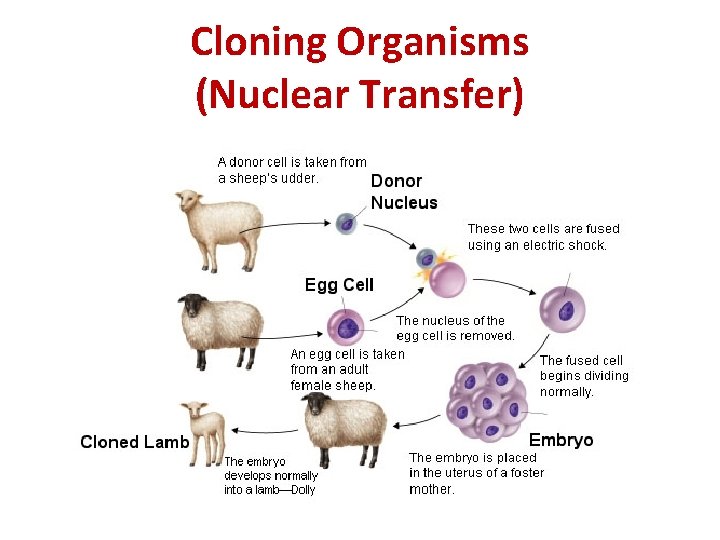
Cloning Organisms • A clone is an organism that is genetically identical to the organism it was produced from (organisms that produce asexually are natural clones of each other) • There are 2 ways to make animal clones: I. Nuclear transfer (the nucleus from a body cell of the organism to be cloned is placed into an egg which has had its nucleus removed. This is then placed into a surrogate mother)

Cloning Organisms (Nuclear Transfer)

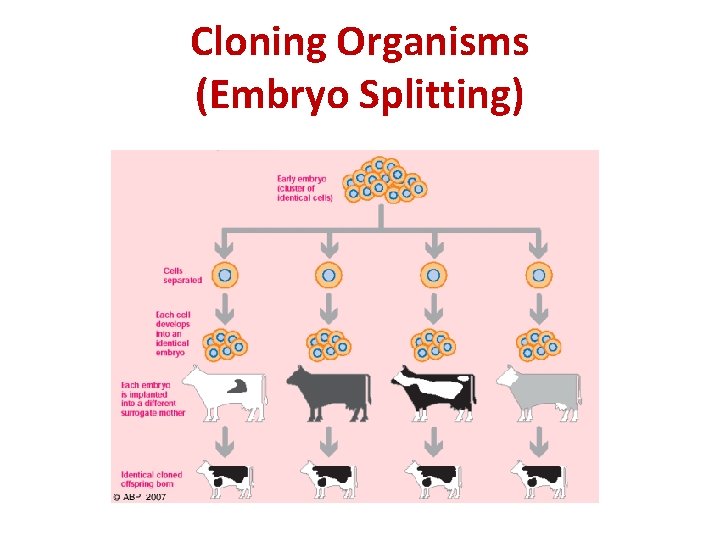
Cloning Organisms (Embryo Splitting) II. Embryo Splitting – cells from the very early stage of an embryo are separated from each other and grown separately into new embryos. These are then placed into different surrogate mothers.

Cloning Organisms (Embryo Splitting)

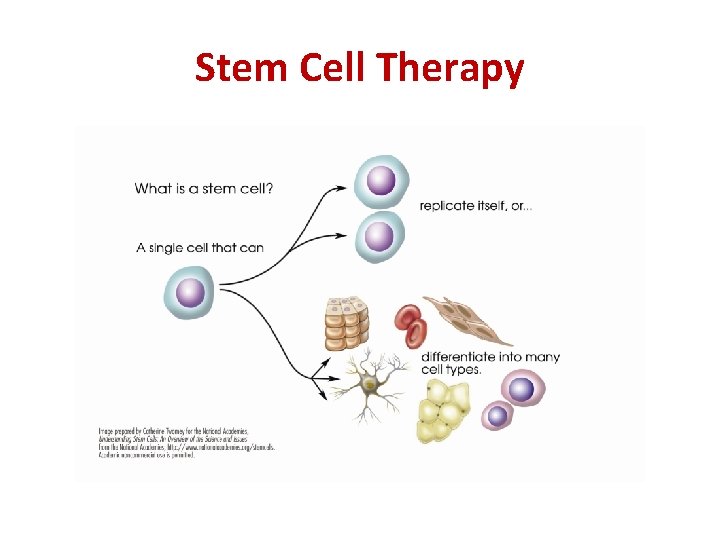
Stem Cell Therapy

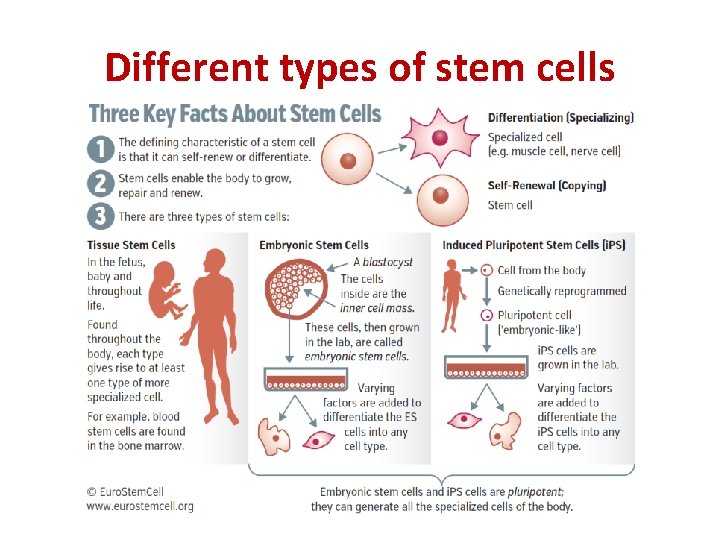
Different types of stem cells

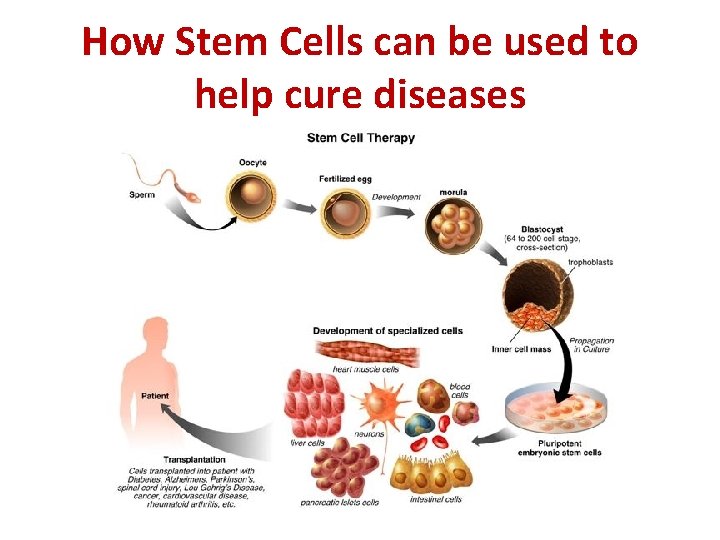
Stem Cell Therapy • Stem cells from an early embryo can be grown in culture (in a petri dish with nutrients) • Different signalling molecules are added to the cells to activate particular genes, allowing them to differentiate into particular types of cells • These healthy cells can be placed into a patient to replace their unhealthy or dying cells

How Stem Cells can be used to help cure diseases

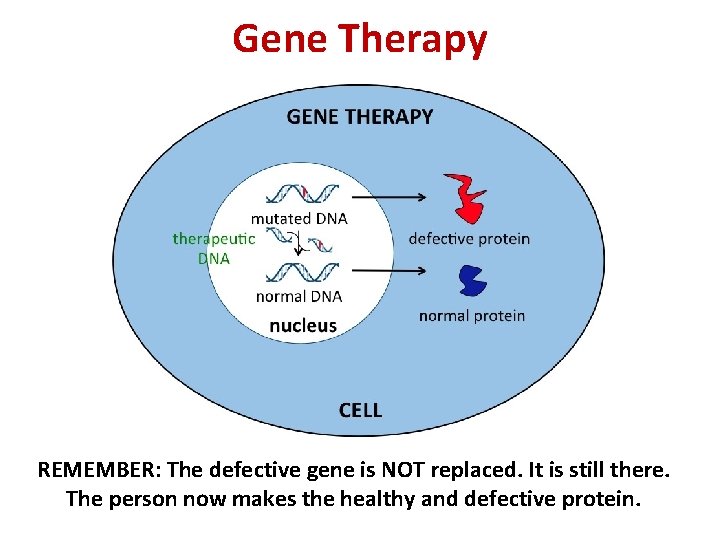
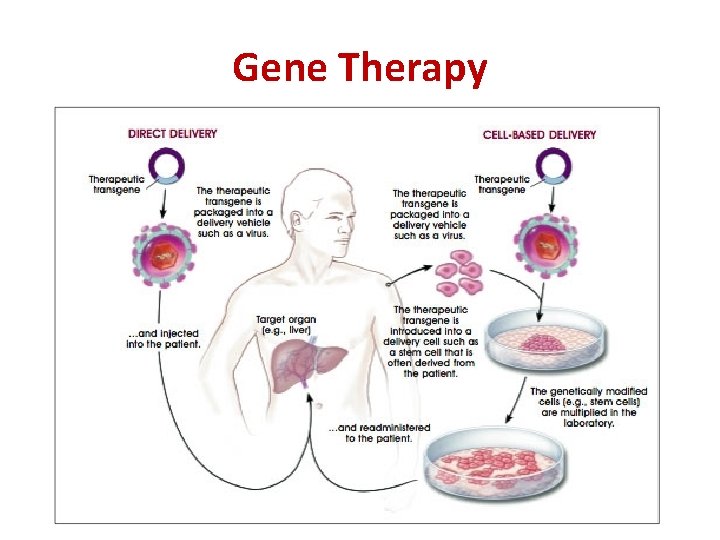
Gene Therapy • Dysfunctional genes in a person may result in essential proteins not being made or not being made properly (e. g. in some forms of blindness or Parkinson’s Disease) • In gene therapy, a vector (usually a virus) is used to deliver a functional copy of the gene to the person’s cells

Gene Therapy REMEMBER: The defective gene is NOT replaced. It is still there. The person now makes the healthy and defective protein.

Gene Therapy