DNA STRUCTURE DNA Structure DNA is a polymer




- Slides: 24

DNA STRUCTURE

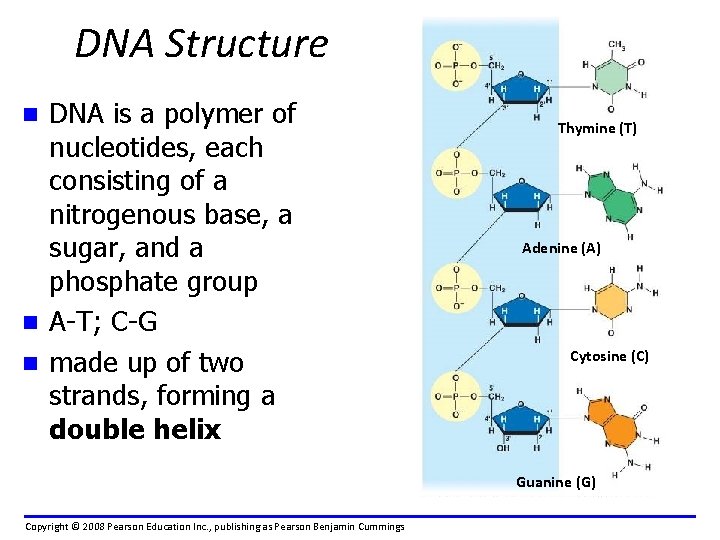
DNA Structure DNA is a polymer of nucleotides, each consisting of a nitrogenous base, a sugar, and a phosphate group n A-T; C-G n made up of two strands, forming a double helix n Thymine (T) Adenine (A) Cytosine (C) Guanine (G) Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

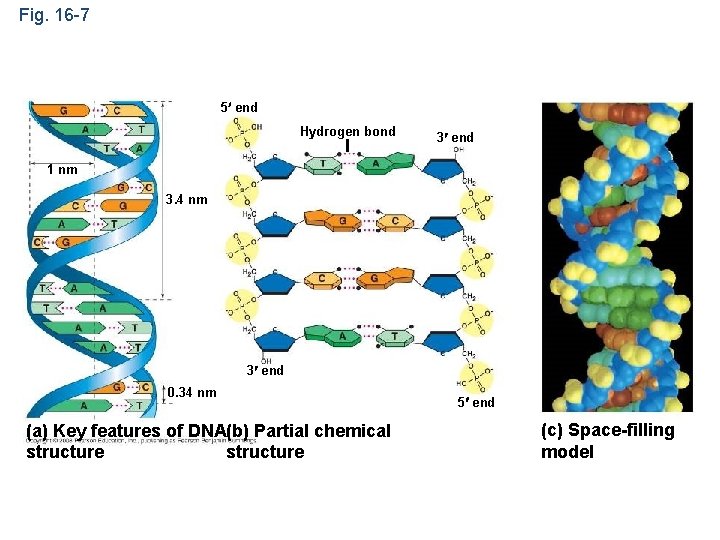
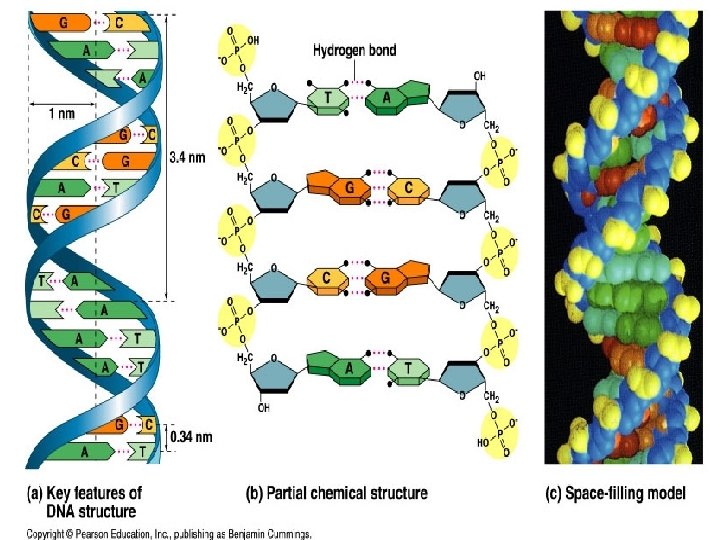
Fig. 16 -7 5 end Hydrogen bond 3 end 1 nm 3. 4 nm 3 end 0. 34 nm (a) Key features of DNA(b) Partial chemical structure 5 end (c) Space-filling model

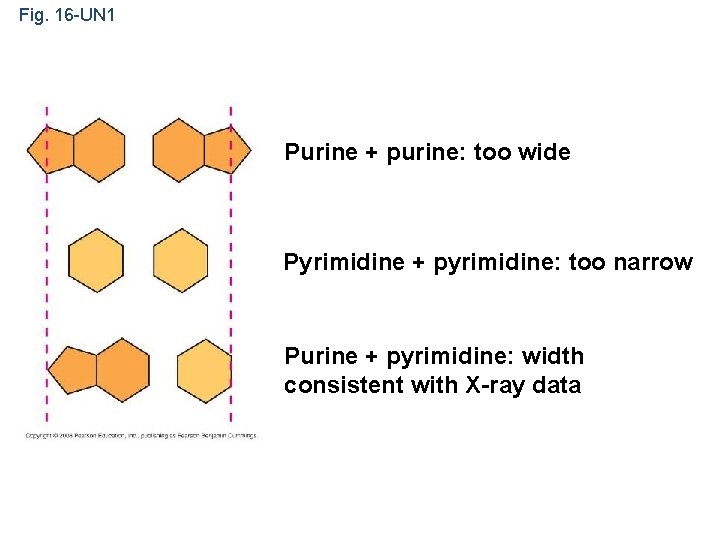
• Watson and Crick built models of a double helix to conform to the X-rays and chemistry of DNA • Franklin had concluded that there were two antiparallel sugar-phosphate backbones, with the nitrogenous bases paired in the molecule’s interior • At first, Watson and Crick thought the bases paired like with like (A with A, and so on), but such pairings did not result in a uniform width • Instead, pairing a purine (2 rings) with a pyrimidine (1 ring) resulted in a uniform width consistent with the X-ray Watson Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

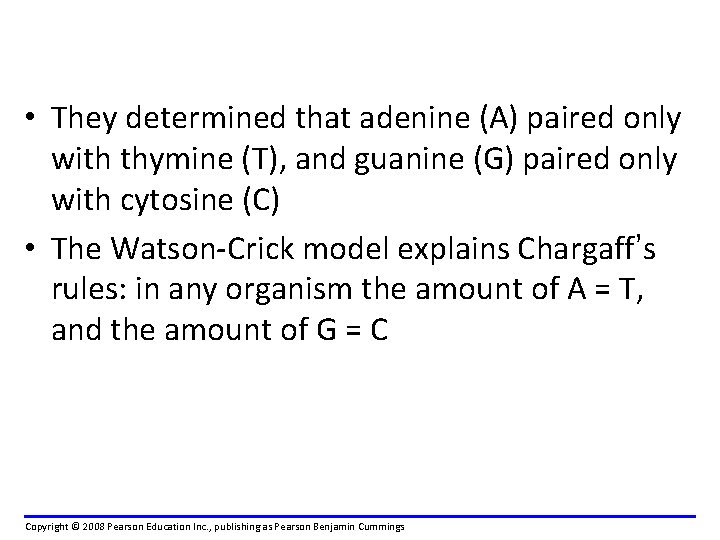
Edwin Chargaff, in 1949, noted two critical bits of data

Fig. 16 -UN 1 Purine + purine: too wide Pyrimidine + pyrimidine: too narrow Purine + pyrimidine: width consistent with X-ray data

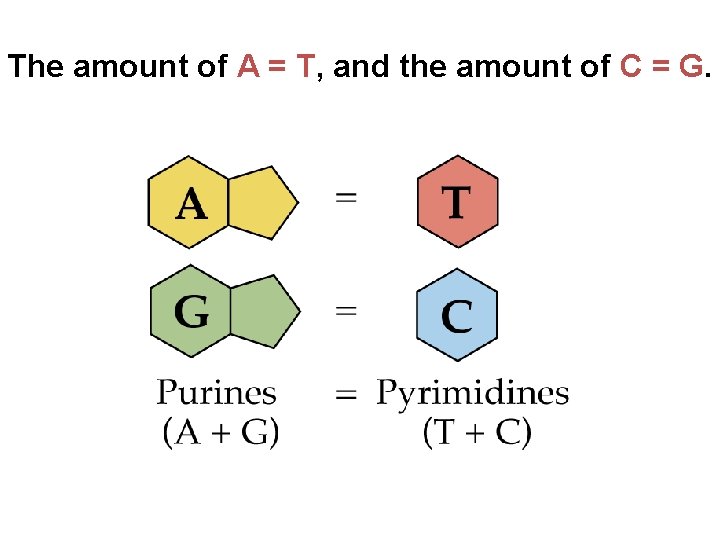
The amount of A = T, and the amount of C = G.

• They determined that adenine (A) paired only with thymine (T), and guanine (G) paired only with cytosine (C) • The Watson-Crick model explains Chargaff’s rules: in any organism the amount of A = T, and the amount of G = C Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

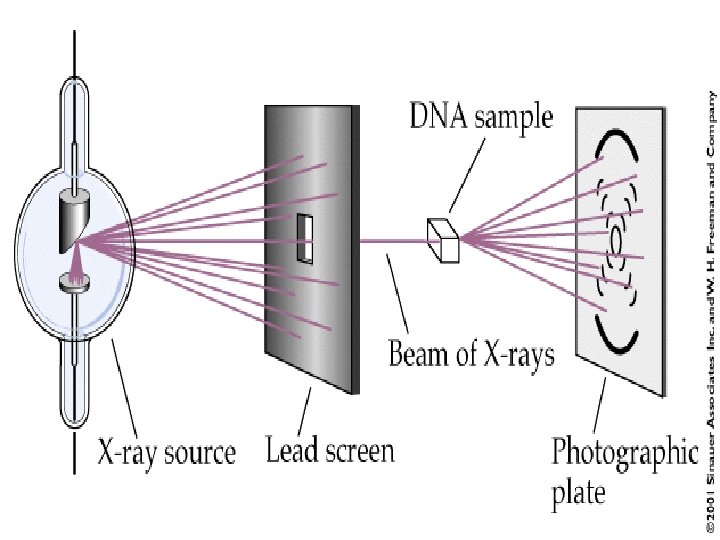
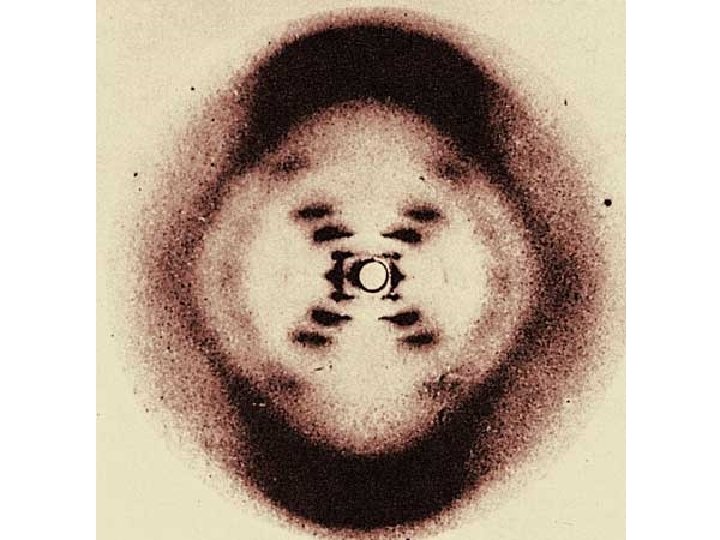
Rosalind Franklin used x-ray diffraction techniques to produce images of DNA molecules. • She concluded: – DNA exists as a long, thin molecule of uniform diameter – – The structure is highly repetitive – – DNA is helical



Watson and Crick used numerous sources of data to build models of DNA.

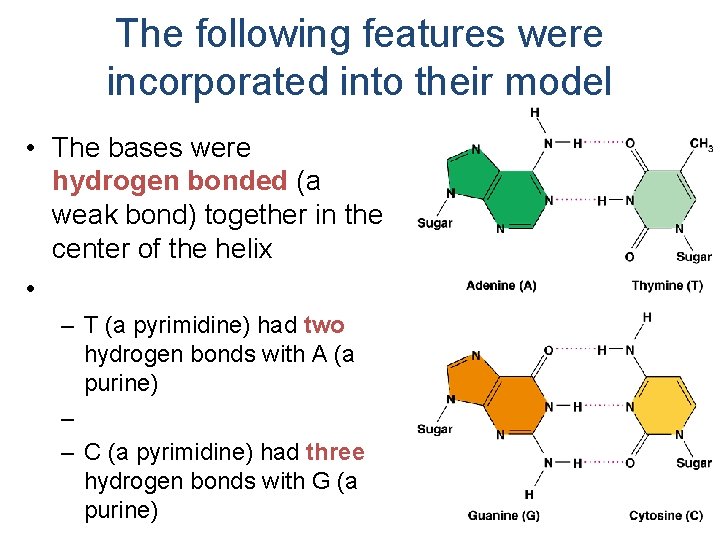
The following features were incorporated into their model • The bases were hydrogen bonded (a weak bond) together in the center of the helix • – T (a pyrimidine) had two hydrogen bonds with A (a purine) – – C (a pyrimidine) had three hydrogen bonds with G (a purine)

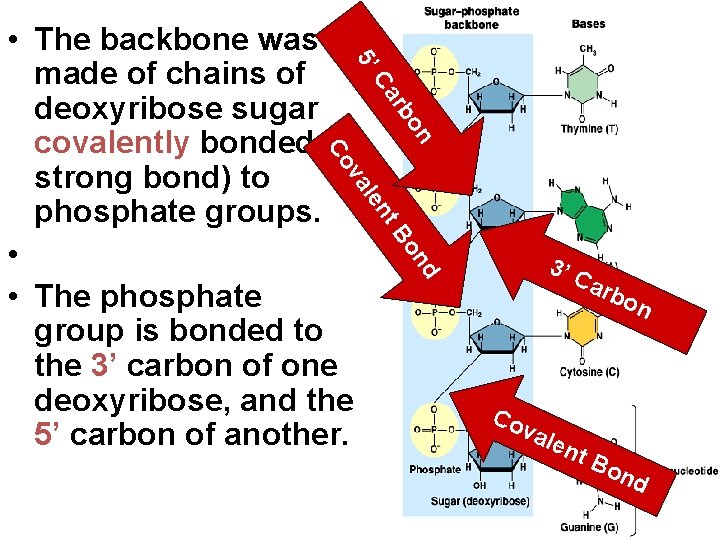
5’ • The backbone was made of chains of deoxyribose sugar covalently bonded (a strong bond) to phosphate groups. • • The phosphate group is bonded to the 3’ carbon of one deoxyribose, and the 5’ carbon of another. n bo r Ca d on t. B en al v Co 3’ C arb Co val ent on Bo nd

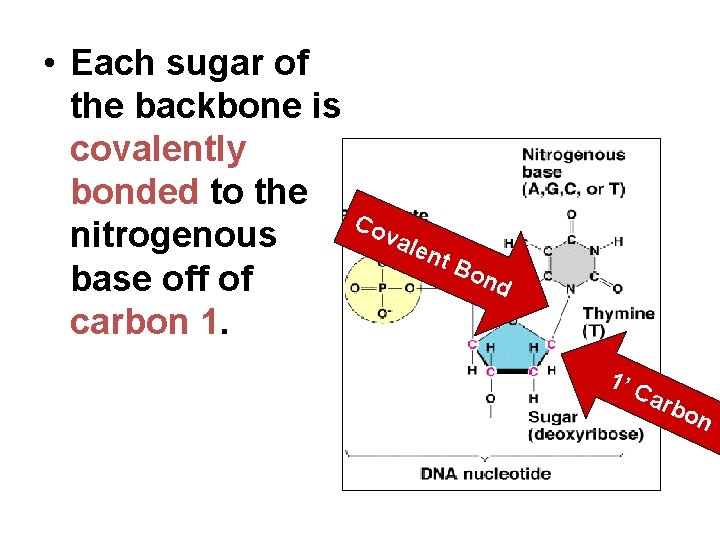
• Each sugar of the backbone is covalently bonded to the nitrogenous base off of carbon 1. Co val ent Bo nd 1’ C arb on

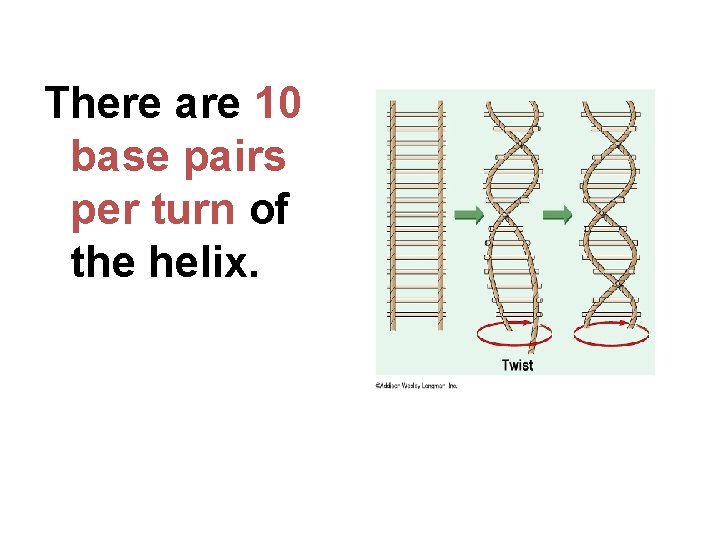
There are 10 base pairs per turn of the helix.

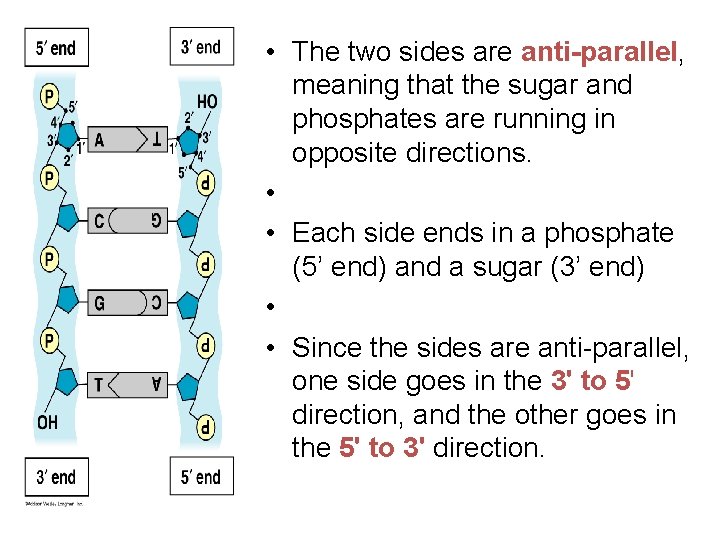
• The two sides are anti-parallel, meaning that the sugar and phosphates are running in opposite directions. • • Each side ends in a phosphate (5’ end) and a sugar (3’ end) • • Since the sides are anti-parallel, one side goes in the 3' to 5' direction, and the other goes in the 5' to 3' direction.


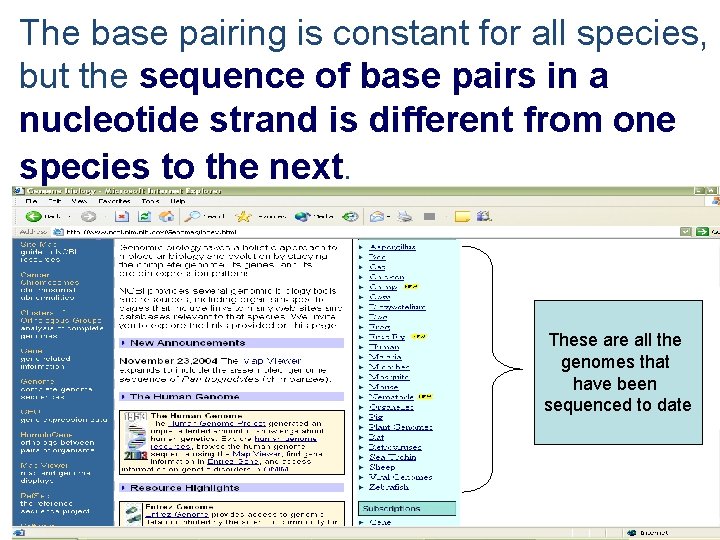
The base pairing is constant for all species, but the sequence of base pairs in a nucleotide strand is different from one species to the next. These are all the genomes that have been sequenced to date

A chromosome consists of a DNA molecule packed together with proteins • The bacterial chromosome is a double-stranded, circular DNA molecule associated with a small amount of protein • Eukaryotic chromosomes have linear DNA molecules associated with a large amount of protein • In a bacterium, the DNA is “supercoiled” and found in a region of the cell called the nucleoid Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

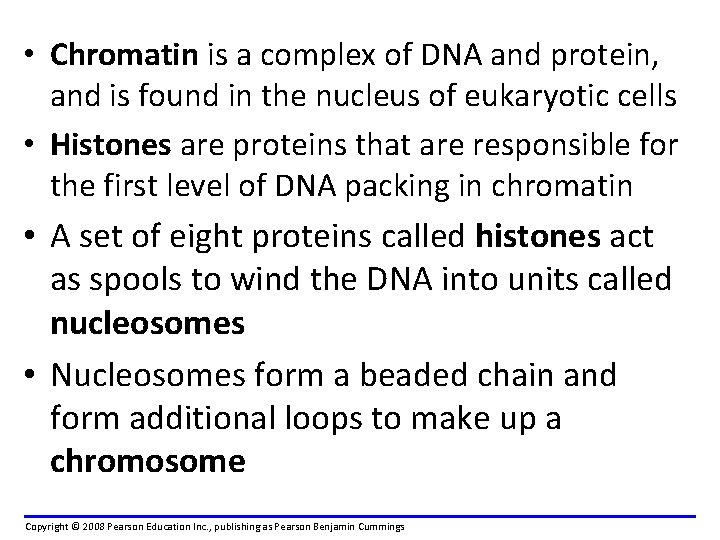
• Chromatin is a complex of DNA and protein, and is found in the nucleus of eukaryotic cells • Histones are proteins that are responsible for the first level of DNA packing in chromatin • A set of eight proteins called histones act as spools to wind the DNA into units called nucleosomes • Nucleosomes form a beaded chain and form additional loops to make up a chromosome Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

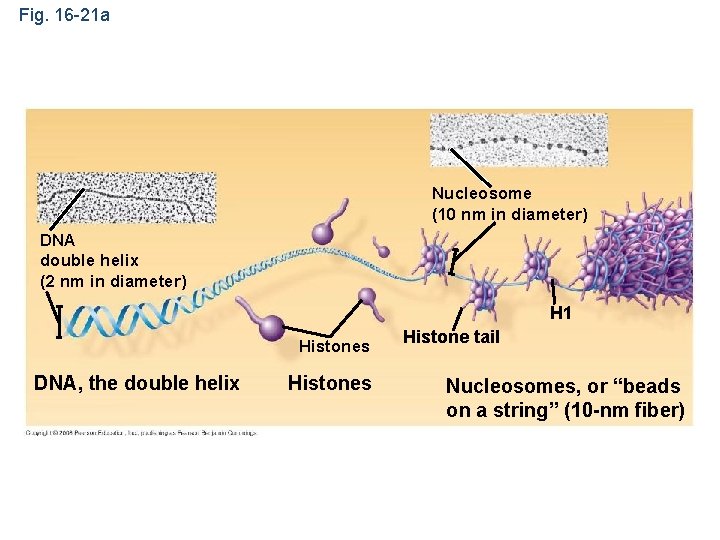
Fig. 16 -21 a Nucleosome (10 nm in diameter) DNA double helix (2 nm in diameter) H 1 Histones DNA, the double helix Histones Histone tail Nucleosomes, or “beads on a string” (10 -nm fiber)

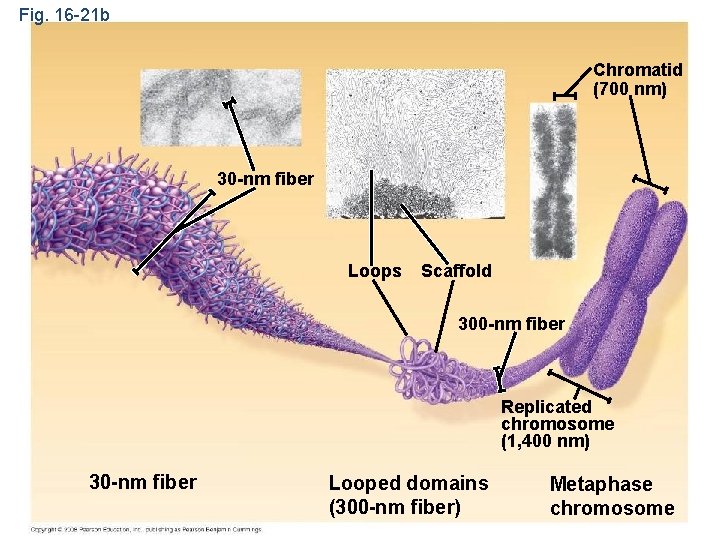
Fig. 16 -21 b Chromatid (700 nm) 30 -nm fiber Loops Scaffold 300 -nm fiber Replicated chromosome (1, 400 nm) 30 -nm fiber Looped domains (300 -nm fiber) Metaphase chromosome

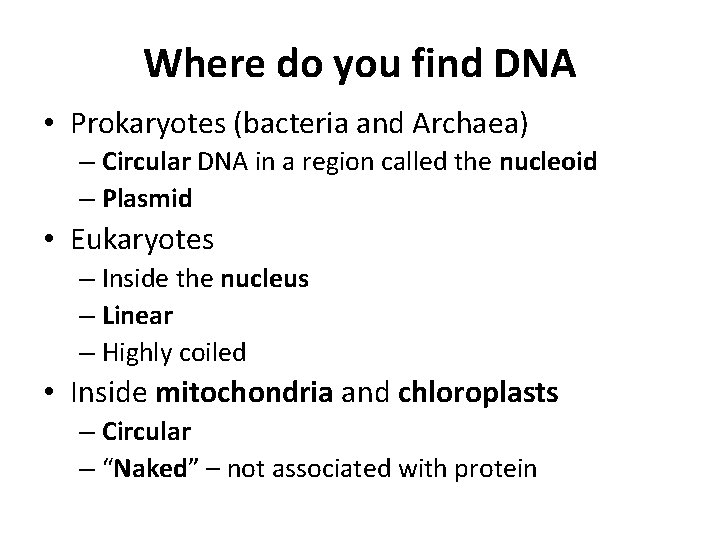
Where do you find DNA • Prokaryotes (bacteria and Archaea) – Circular DNA in a region called the nucleoid – Plasmid • Eukaryotes – Inside the nucleus – Linear – Highly coiled • Inside mitochondria and chloroplasts – Circular – “Naked” – not associated with protein