DNA structure DNA is a polymer made of















































- Slides: 67

DNA structure

DNA is a polymer made of four monomers (A, T, G, C). Each monomer is contains: a. a nucleobase (nitrogeneous base) attached to a sugar (deoxyribose) and a phosphate. b. a nucleobase (nitrogeneous base) attached to a sugar (deoxyribose) and a lipid c. a nucleobase (nitrogeneous base) attached to an amino acid and a phosphate. d. an amino acid attached to a sugar (deoxyribose) and a phosphate.

True or False: One end of a DNA strand is called the 5’-end and the other end is called the 3’-end. The two strands of DNA are anti-parallel, meaning they run in opposite directions. The backbone of DNA (the strands) are composed of sugars (deoxyribose) attached to each by phosphates.

If you know the sequence of one strand, you can figure out the sequence of the other. If the sequence of one strand is 5’-ATGCAT-3’, what is the sequence of its complementary strand?

The two strands of DNA are held together by Hydrogen bonds between the nucleobases. There are three hydrogen bonds holding together G: : : C, and two hydrogen bonds holding together A: : T. Which of the following strands of DNA is most stable? 1. A. 5’GCGGCC-3’ 2. 3’CGCCGG-5’ 3. B. 5’AAAAAA-3’ 4. 3’TTTTTT-5’ 5. C. 5’TATATG-3’ 6. 3’ATATAC-5’ 7. D. 5’ATGCAT-3’ 8. 3’TACGTA-5’

DNA replication sdfds

What enzyme unzips (unwinds) the DNA molecule? a. b. c. d. e. DNA polymerase Helicase Primase DNA ligase unzipase

Which of the following are limitations of DNA polymerase? a. It can only add bases to the exposed 3’ end of a preexisting strand b. It can only replicate the leading strand c. It can only replicate the lagging strand d. None of the above

T or F: The point where separation of the DNA occurs is called the replication fork.

Put the following events in the proper order: a. DNA polymerase extends the RNA primer in opposite directions using monomers present in the cell. b. DNA ligase joins together two adjacent strands of DNA c. Helicase unwinds/separates the two DNA strands c. RNA primase then adds a short complementary strand of RNA (a RNA primer) to each strand d. DNA polymerase extends each strand until it runs into another section that is already copied

Bidirectional DNA replication

DNA replication in bacteria begins at: a. b. c. d. e. A single origin and proceeds in one direction A single origin and proceeds in both directions Two origins and proceeds in both directions Many origins and proceeds in one direction Many origins and proceeds in two directions

True or False: The bacterial chromosome is linear

m. RNA synthesis (transcription) Can you find a mistake in this animation?

The segment of the DNA molecule where m. RNA synthesis begins is called the: a. b. c. d. e. Promoter region Sigma factor Transcription terminator Polymerase Template

The complementary m. RNA stand that would be synthesized from the DNA sequence 5’-CTGAC-3’ would be: a. b. c. d. 3’-GACTG-5’ 3’-GACUG-5’ 3’-CTGAC-5’ 5’-CTGAC-3’

The name of the structure that causes the synthesis of m. RNA to cease is called the: a. b. c. d. e. Promoter region Sigma factor Transcription terminator Polymerase Template

T or F: The synthesis of m. RNA is called translation. The synthesis of m. RNA is in the 5’ to 3’ direction. The name of the enzyme that synthesizes m. RNA is RNA polymerase

Exon Shuffling:

What is the name of the part of m. RNA (in the primary transcript) that does not encode a protein, and is removed by splicing before a protein is made: a. Exon b. Intron c. Gene d. 5’cap e. 3’poly. A tail

The 5’-cap and 3’-poly. A tails added to m. RNA serve what purpose? a. to make the m. RNA more stable b. to make the m. RNA longer c. to enable splicing out of introns d. All of the above

One m. RNA can be spliced many different ways, including all or just some of the exons. This allows 30, 000 genes in humans to make over 100, 000 different proteins. This is called: a. Alternative splicing b. Intron shuffling c. Transcription d. translation

Processing of Gene Information: Prokaryotes vs Eukaryotes

Which of the following statements is true for eukaryotic m. RNA, but not true for prokaryotic m. RNA? a. A cap is added to their 5’end. b. A poly-A tail is added to their 3’ end. c. Each usually specifies only a single protein. d. All of the above are true for eukaryotic m. RNA, and false for prokaryotic m. RNA.

T or F: Prokaryotic m. RNA can be translated by many ribosomes at the same time as it is being transcribed from DNA, but eukaryotic m. RNA must first be processed and then transported from nucleus to the cytoplasm before it can be translated into a protein. An operon consists of a set of related genes that are transcribed as a single m. RNA and are found in eukaryotes.

Transposons: aka jumping genes Does anyone know what a bacterial plasmid is?

Transposons are: a. Segments of m. RNA b. Plasmids c. Segments of DNA d. Segments of transfer RNA (t. RNA)

Transposons are transported between different cells by: a. Transduction b. A sex pilus c. Conjugation d. Bacteriophages e. Plasmids

Which of the following is true about transposons (include all that are true)? a. They are capable of moving from one cell to another b. They can move into the host cell genome c. They can more from one site in the host cell genome to another site d. They can replicate themselves before moving

Addition and deletion mutations

The nucleic acid sequence in m. RNA is determined by: a. b. c. d. The order of amino acids in the protein The nucleotide sequence in DNA The nucleotide sequence in t. RNA All of the above

T or F: One amino acid in a protein chain is encoded by three consecutive bases in RNA; the three bases in m. RNA that code for one amino acid is called a codon.

Classify the following three “mutations” as either a point mutation or a frame-shift mutation. If it is a frame-shift mutation, is it an addition or deletion? Original mutant#1 mutant#2 mutant#3 THE BIG FAT CAT ATE ALL THE ICE THE BIG ATC ATA TEA LLT HEI CEN THE BIG FAT CAT TAT EAL LTH EIC THE BIG CAT ATE ALL THE ICE What type of mutation has the least impact on the message?

If a frame-shift mutation creates a stop codon in the DNA sequence, then a. b. c. d. The resulting protein will not be affected The phenotype will change but not the genotype The resulting protein will be too short and non-functional The resulting protein will be too long and non-functional

A nucleotide deletion that occurs during DNA replication: a. b. c. d. e. Causes one amino acid of the protein to be deleted Causes all of the amino acids of the protein to be incorrect Causes the amino acids after the deletion to be corrected Causes the amino acids before the deletion to be incorrect Has no effect on the resulting protein

Mutations in DNA, such as point mutations or frame-shift mutations, can be caused by: a. b. c. d. e. Uncorrected errors during DNA replication Mutagens such as the chemicals found in smoke High energy particles such as radiation and UV light Insertion of transposons into an exonic region All of the above

Restriction Endonucleases

Which of the following could NOT be the recognition site of a restriction endonuclease? a. GAATTC CTTAAG b. ATCGAT TAGCTA c. CTGCAG GACGTC d. GCTTGC CGAACG e. GGATCC CCTAGG

The single-stranded ends of DNA molecules can be joined together by: a. b. c. d. e. Restriction endonucleases DNA ligase DNA polymerase Primase Helicase

Human DNA cut with restriction enzyme A will have the same sticky ends as (and can therefore be ligated/joined to) which of the following: a. b. c. d. e. Human DNA cut with restriction enzyme B Human DNA that is uncut Bacterial DNA that is uncut Corn DNA that is cut with restriction enzyme A None of the above

T or F: The ligating (joining) of sticky ends involves the formation of phosphodiester bonds.

If you have a piece of bacterial DNA that is 1280 bases long, and you add to it a restriction endonuclease that recognizes and cuts the following sequence: 5’-CATG-3’ 3’-GTAC-5’ approximately how many times will the bacterial DNA be cut? Hint: if you flip a coin four times, how often to you expect to get heads four times in a row?

PCR: Polymerase Chain Reaction

PCR requires all of the following except: a. b. c. d. e. Primers DNA ligase DNA polymerase (usually Taq DNA polymerase) At least one strand of DNA (polymer) to copy/amplify Deoxyribonucleotides (monomers)

The purpose of heating the DNA during each cycle of PCR is to a. b. c. d. Activate the polymerase enzyme Speed up catalysis by the polymerase enzyme Separate the two strands of DNA template Increase binding of primers to the DNA template

T or F: Even though Taq polymerase is an enzyme from a bacteria, it can copy DNA from any species, including humans. During the PCR, the hydrogen bonds holding the two strands of DNA molecules together are broken by the helicase enzyme.

You can determine which part of OJ Simpon’s DNA you wish to amplify by changing which of the following: a. The sequence of the primer b. The temperature of the PCR c. The amount of polymerase

If you start with one drop of OJ Simpson’s blood, containing ten cells (and therefore ten copies of his DNA), and you amplify it by PCR for 10 cycles of “heat-cool-copy-repeat”, how many copies of the DNA have you made?

What temperature do you think your DNA polymerase works best at? Do you think your DNA polymerase works at 95 C (nearly the temperature of boiling water)? Where do you think scientists found a polymerase that is active even at very high temperatures, such as Taq DNA polymerase?

Reconstruction of the Primate Alcohol Metabolic Pathway: Did Ardi Party Hearty? Matthew Carrigan Research Fellow Foundation for Applied Molecular Evolution www. ffame. org

Ethanol Metabolism Ethanol can be a valuable source of energy NADH + +

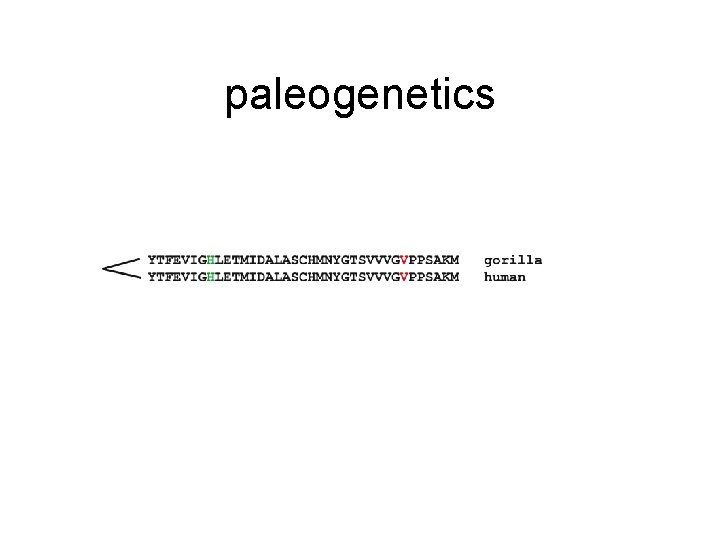
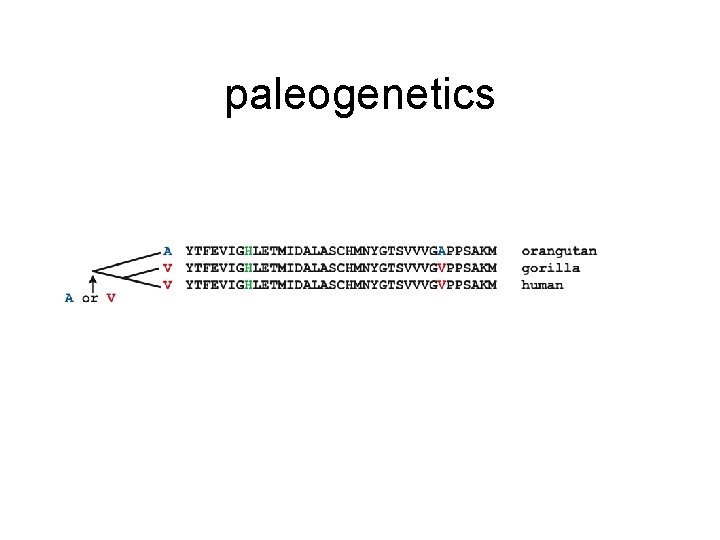
paleogenetics

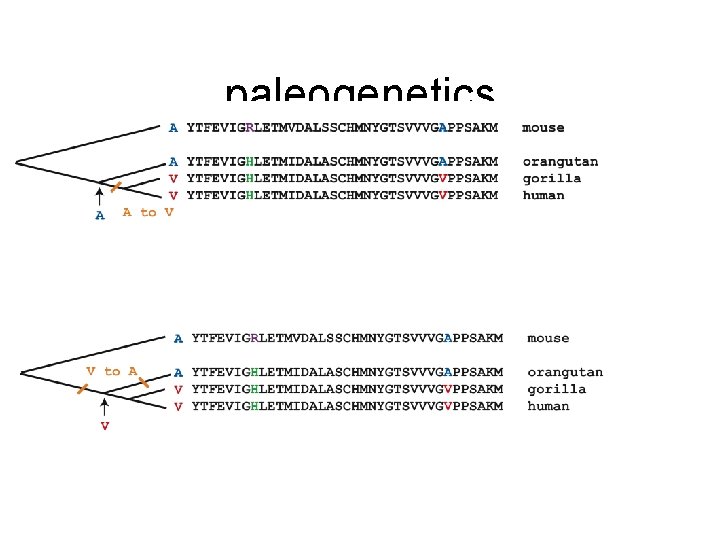
paleogenetics

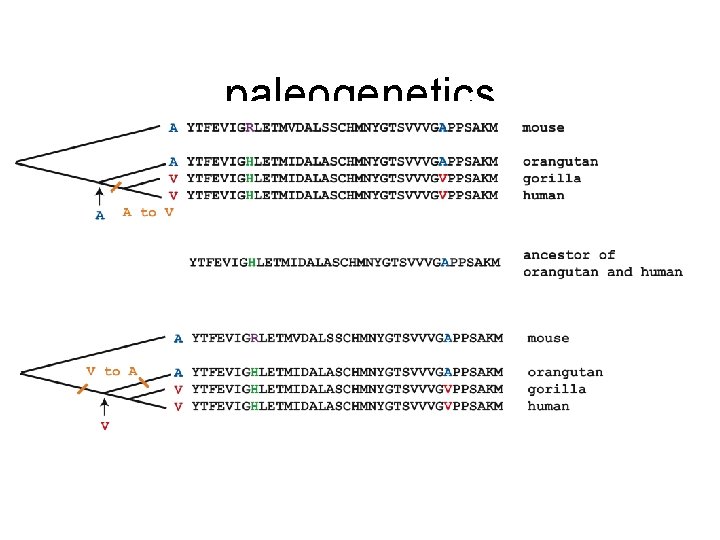
paleogenetics

paleogenetics

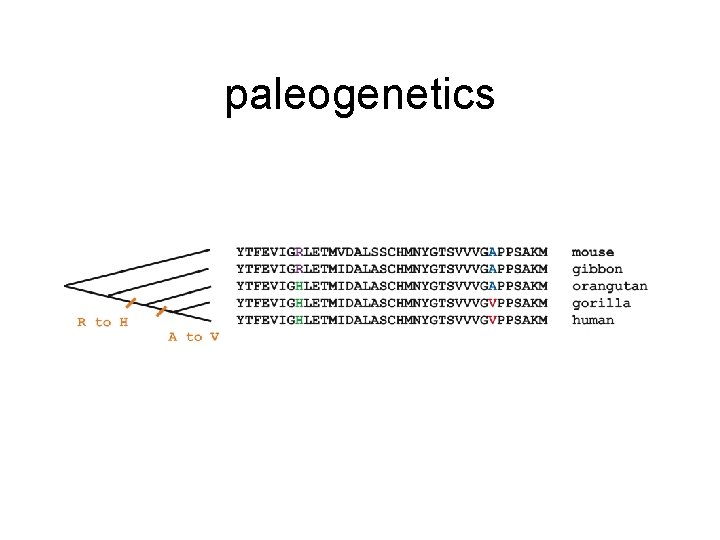
paleogenetics

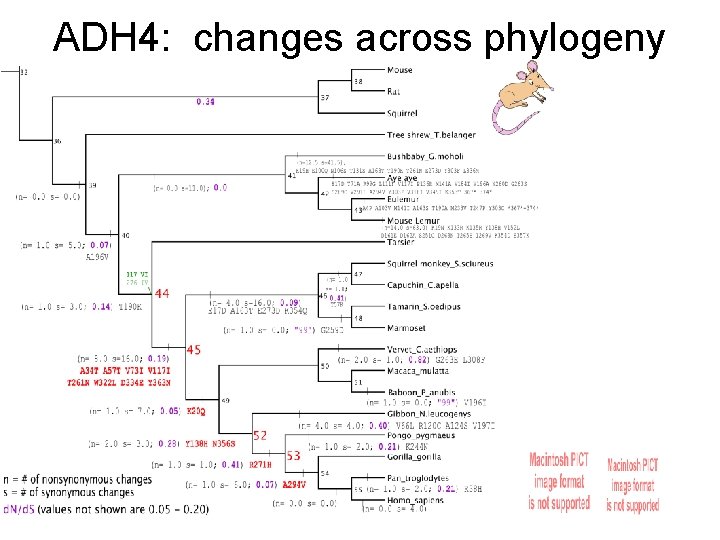
ADH 4: changes across phylogeny

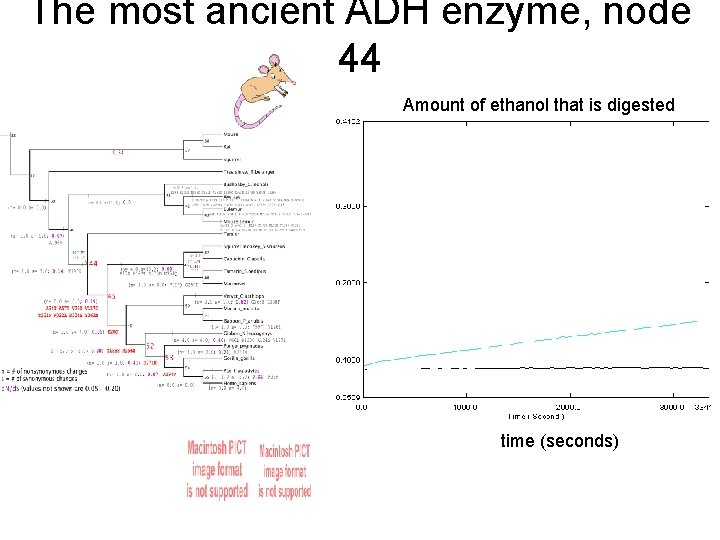
The most ancient ADH enzyme, node 44 Amount of ethanol that is digested time (seconds)

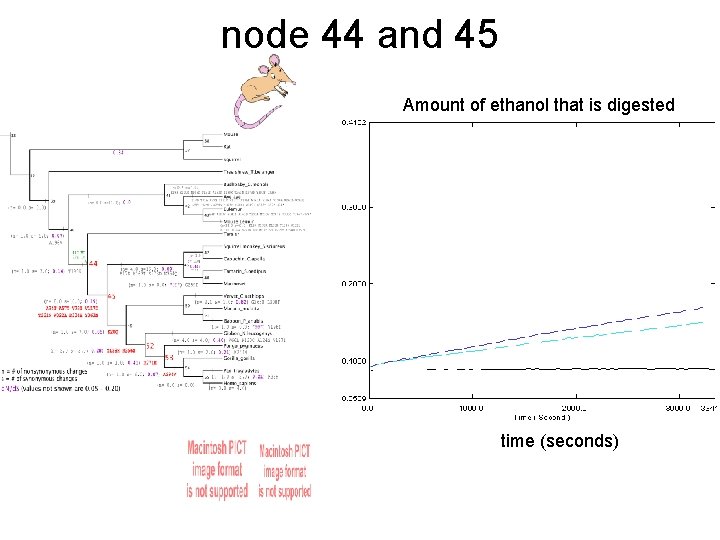
node 44 and 45 Amount of ethanol that is digested time (seconds)

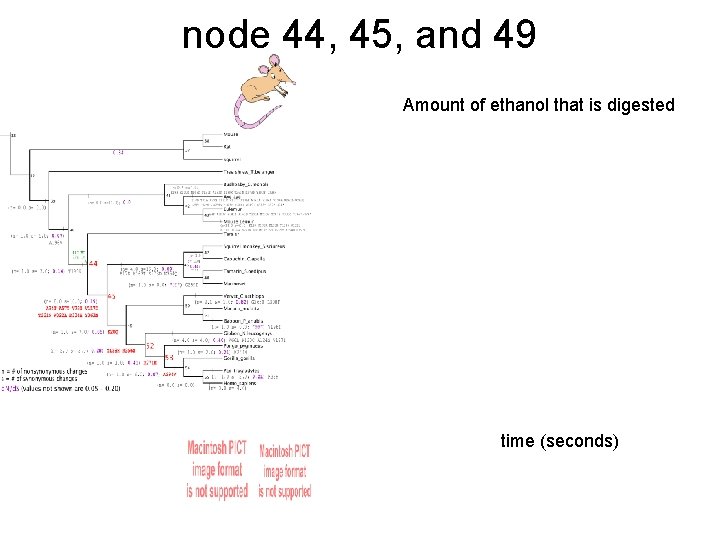
node 44, 45, and 49 Amount of ethanol that is digested time (seconds)

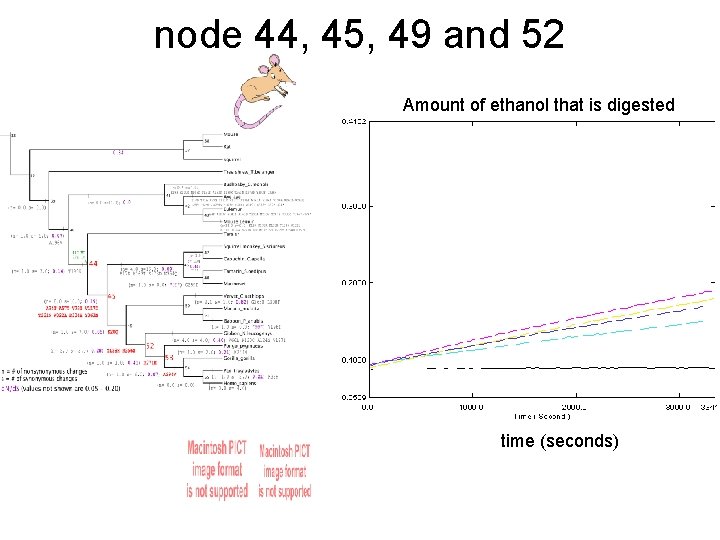
node 44, 45, 49 and 52 Amount of ethanol that is digested time (seconds)

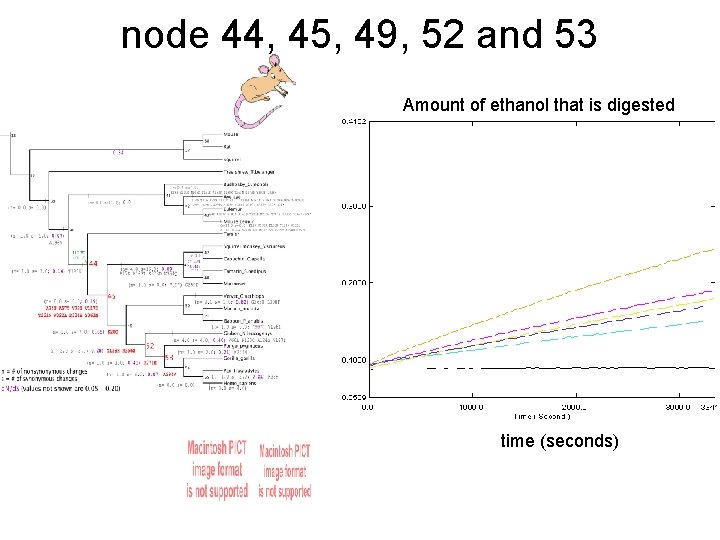
node 44, 45, 49, 52 and 53 Amount of ethanol that is digested time (seconds)

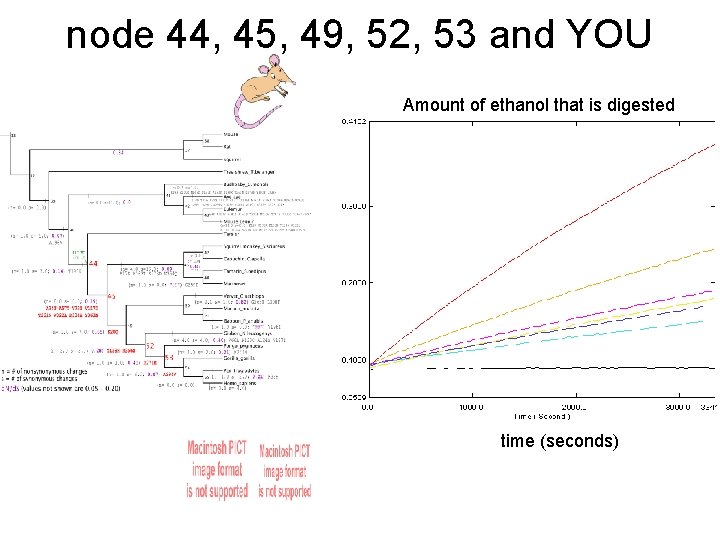
node 44, 45, 49, 52, 53 and YOU Amount of ethanol that is digested time (seconds)

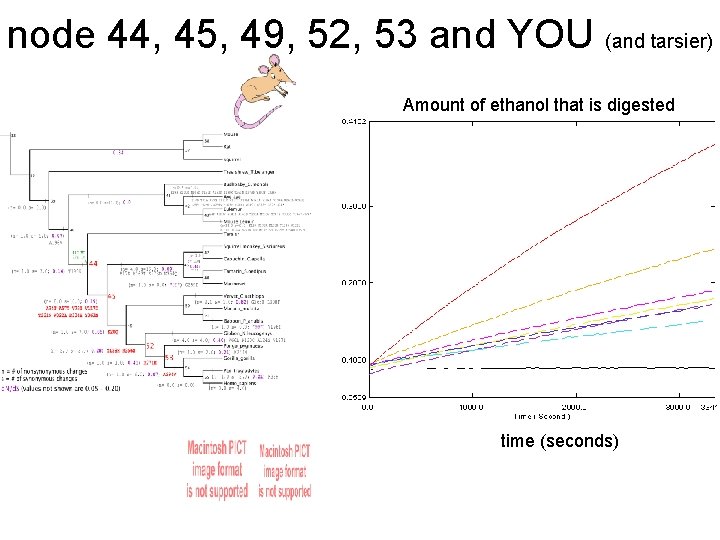
node 44, 45, 49, 52, 53 and YOU (and tarsier) Amount of ethanol that is digested time (seconds)

So are you saying we are adapted to drink ethanol?

Ethanol Metabolism Ethanol can be a valuable source of energy… …but if you have too much, a toxic intermediate can accumulate NADH + +

…and if we (and orangutans) are adapted to drink ethanol… then where all the drunk monkeys?