Support Vector Machine Data Mining Olvi L Mangasarian



- Slides: 28

Support Vector Machine Data Mining Olvi L. Mangasarian with Glenn M. Fung, Jude W. Shavlik & Collaborators at Exon. Hit – Paris Data Mining Institute University of Wisconsin - Madison

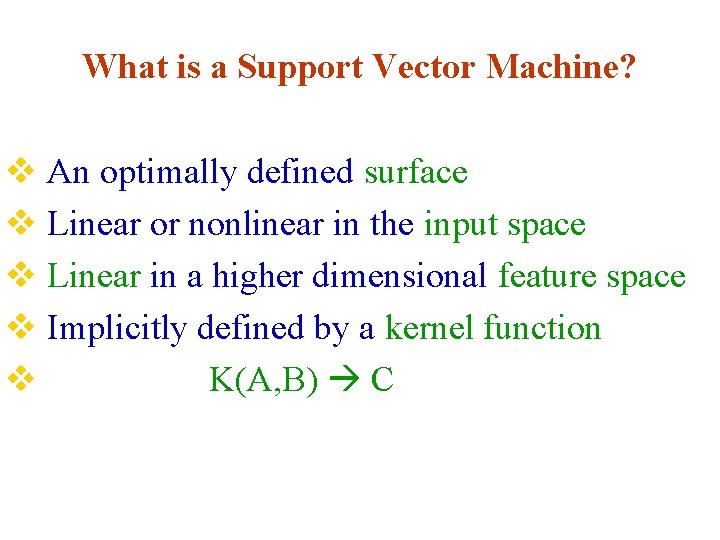
What is a Support Vector Machine? v An optimally defined surface v Linear or nonlinear in the input space v Linear in a higher dimensional feature space v Implicitly defined by a kernel function v K(A, B) C

What are Support Vector Machines Used For? v Classification v Regression & Data Fitting v Supervised & Unsupervised Learning

Principal Topics v. Knowledge-based classification ØIncorporate expert knowledge into a classifier v. Breast cancer prognosis & chemotherapy ØClassify patients on basis of distinct survival curves Ø Isolate a class of patients that may benefit from chemotherapy v. Multiple Myeloma detection via gene expression measurements v. Drug discovery based on gene macroarray expression v. Joint work with Exon. Hit

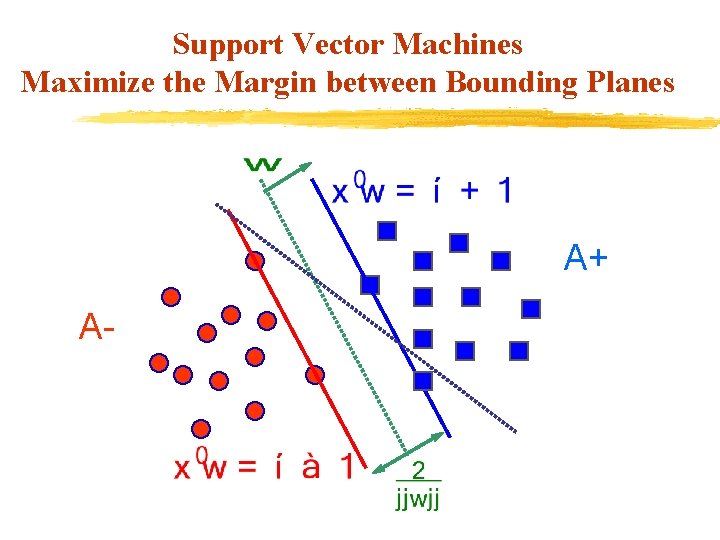
Support Vector Machines Maximize the Margin between Bounding Planes A+ A-

Principal Topics v. Knowledge-based classification (NIPS*2002)

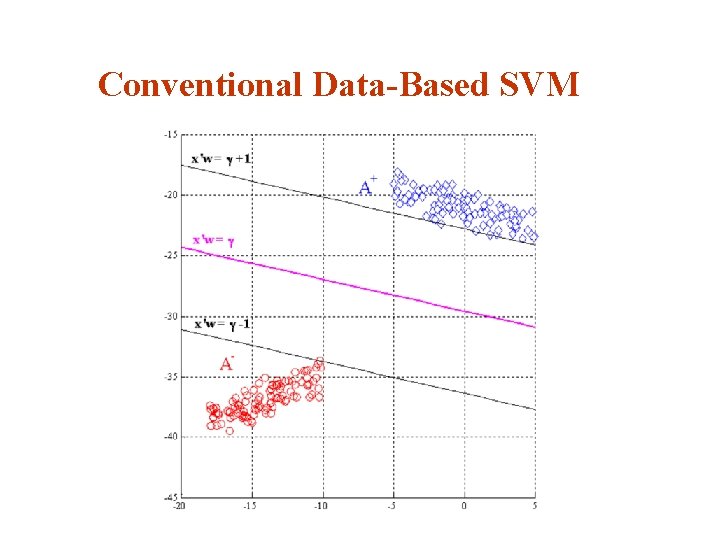
Conventional Data-Based SVM

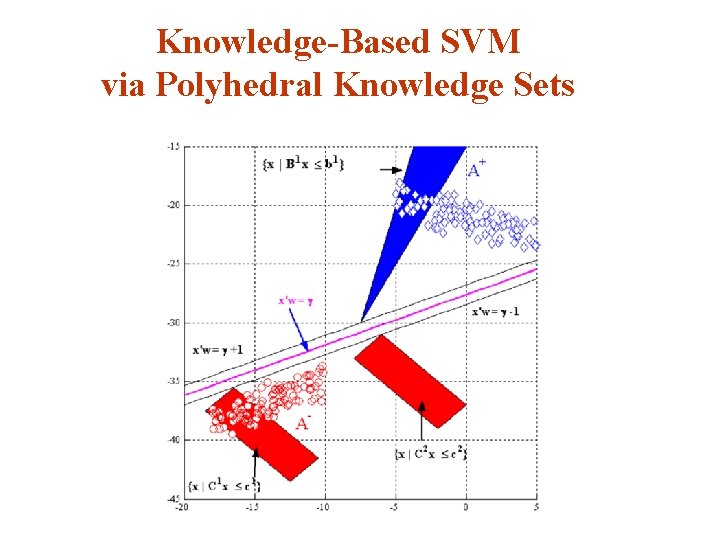
Knowledge-Based SVM via Polyhedral Knowledge Sets

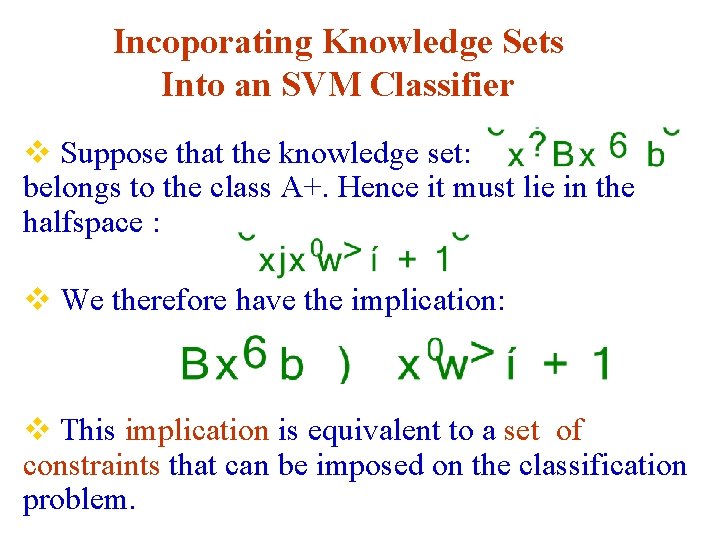
Incoporating Knowledge Sets Into an SVM Classifier v Suppose that the knowledge set: belongs to the class A+. Hence it must lie in the halfspace : v We therefore have the implication: v This implication is equivalent to a set of constraints that can be imposed on the classification problem.

Numerical Testing The Promoter Recognition Dataset v Promoter: Short DNA sequence that precedes a gene sequence. v A promoter consists of 57 consecutive DNA nucleotides belonging to {A, G, C, T}. v Important to distinguish between promoters and nonpromoters v This distinction identifies starting locations of genes in long uncharacterized DNA sequences.

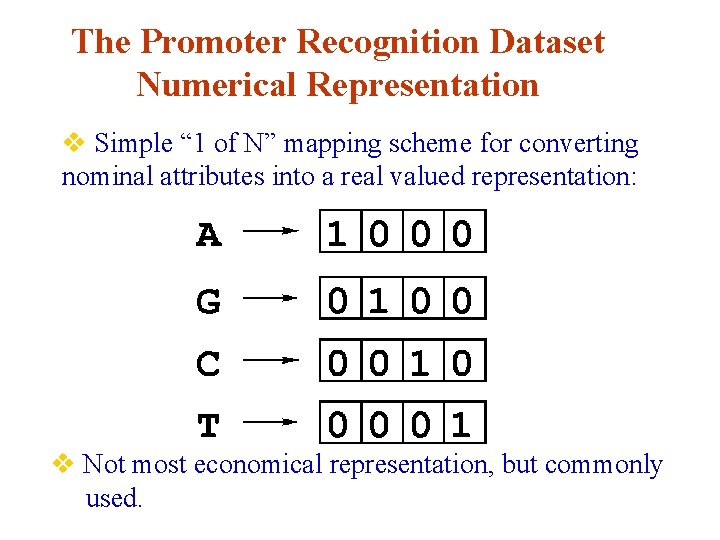
The Promoter Recognition Dataset Numerical Representation v Simple “ 1 of N” mapping scheme for converting nominal attributes into a real valued representation: v Not most economical representation, but commonly used.

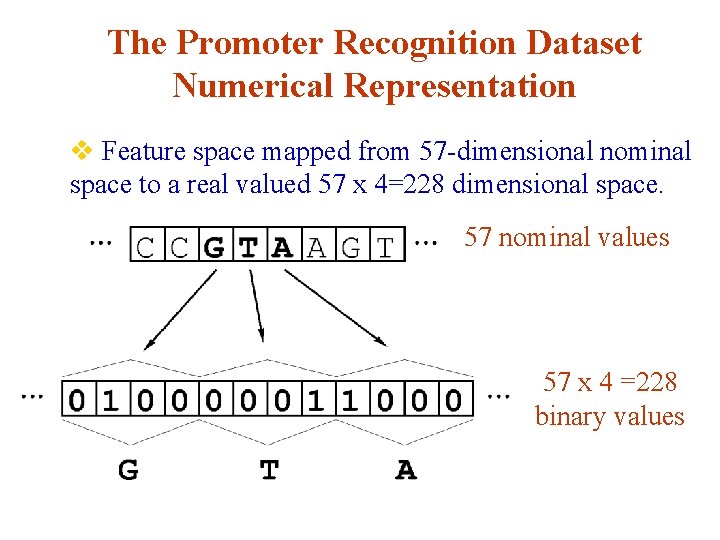
The Promoter Recognition Dataset Numerical Representation v Feature space mapped from 57 -dimensional nominal space to a real valued 57 x 4=228 dimensional space. 57 nominal values 57 x 4 =228 binary values

Promoter Recognition Dataset Prior Knowledge Rules v Prior knowledge consist of the following 64 rules:

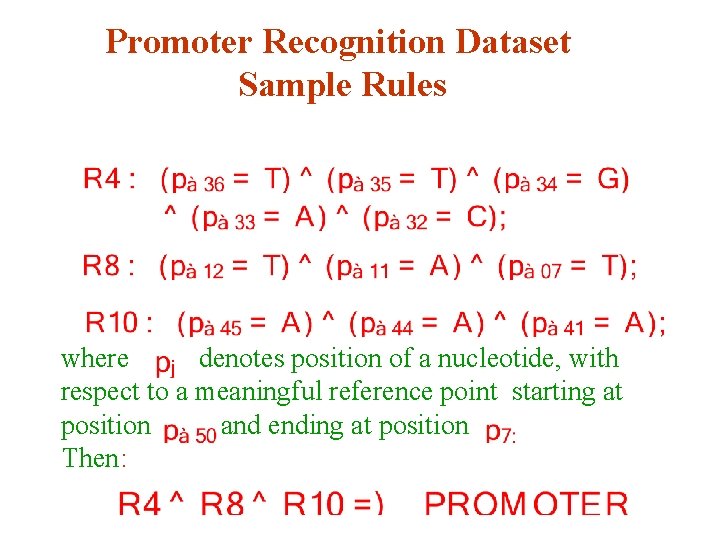
Promoter Recognition Dataset Sample Rules where denotes position of a nucleotide, with respect to a meaningful reference point starting at position and ending at position Then:

The Promoter Recognition Dataset Comparative Algorithms v KBANN Knowledge-based artificial neural network [Shavlik et al] v BP: Standard back propagation for neural networks [Rumelhart et al] v O’Neill’s Method Empirical method suggested by biologist O’Neill [O’Neill] v NN: Nearest neighbor with k=3 [Cost et al] v ID 3: Quinlan’s decision tree builder[Quinlan] v SVM 1: Standard 1 -norm SVM [Bradley et al]

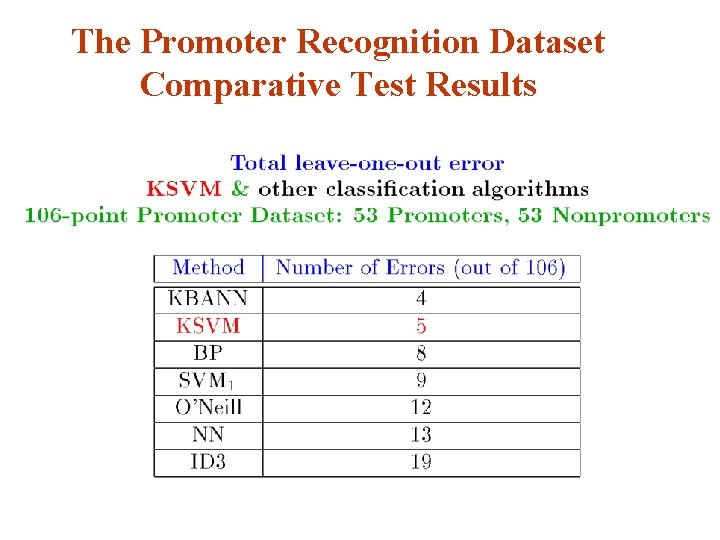
The Promoter Recognition Dataset Comparative Test Results

Principal Topics v. Breast cancer prognosis & chemotherapy

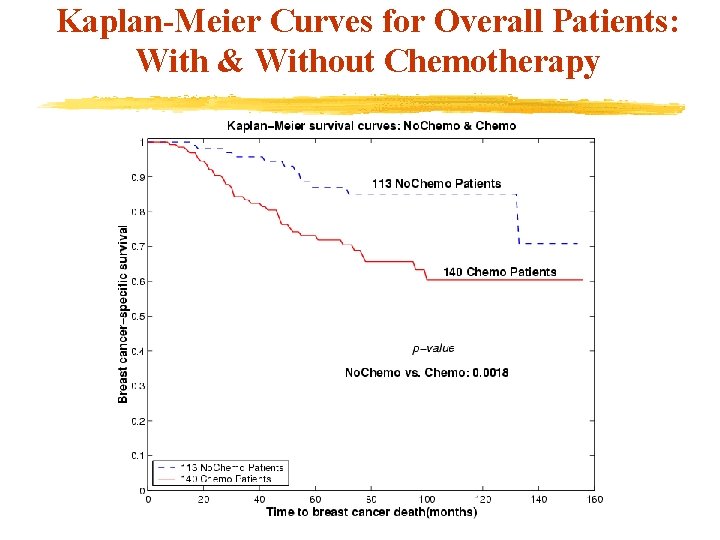
Kaplan-Meier Curves for Overall Patients: With & Without Chemotherapy

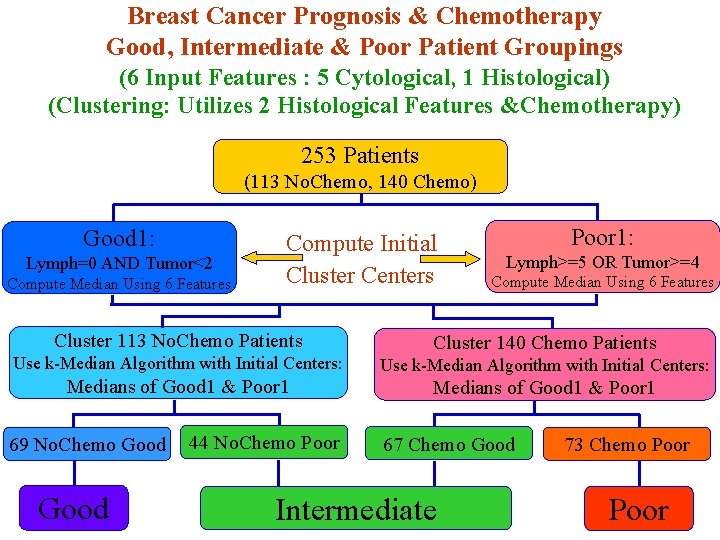
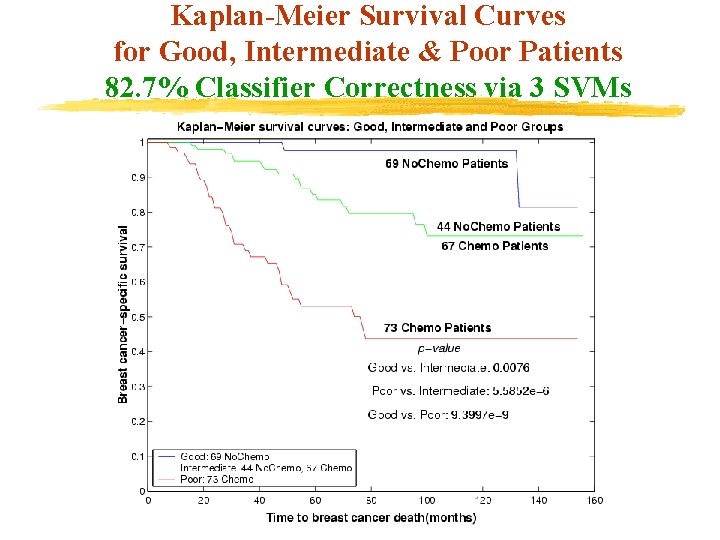
Breast Cancer Prognosis & Chemotherapy Good, Intermediate & Poor Patient Groupings (6 Input Features : 5 Cytological, 1 Histological) (Clustering: Utilizes 2 Histological Features &Chemotherapy) 253 Patients (113 No. Chemo, 140 Chemo) Good 1: Lymph=0 AND Tumor<2 Compute Median Using 6 Features Compute Initial Cluster Centers Poor 1: Lymph>=5 OR Tumor>=4 Compute Median Using 6 Features Cluster 113 No. Chemo Patients Cluster 140 Chemo Patients Use k-Median Algorithm with Initial Centers: Medians of Good 1 & Poor 1 69 No. Chemo Good 44 No. Chemo Poor 67 Chemo Good Intermediate 73 Chemo Poor

Kaplan-Meier Survival Curves for Good, Intermediate & Poor Patients 82. 7% Classifier Correctness via 3 SVMs

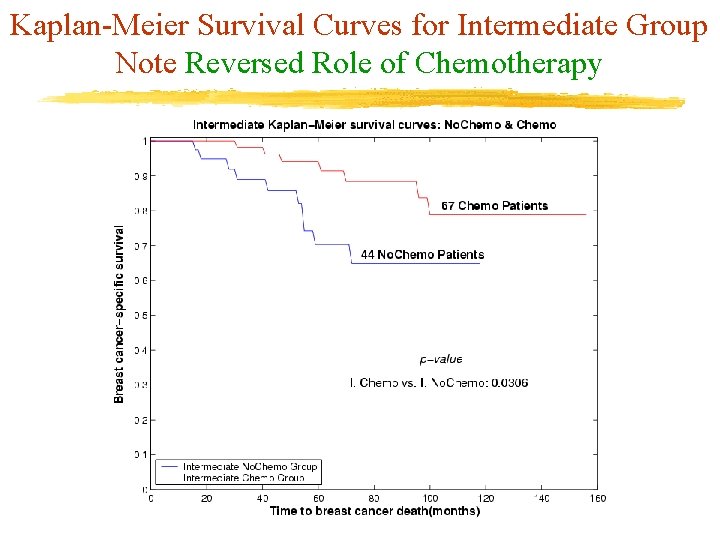
Kaplan-Meier Survival Curves for Intermediate Group Note Reversed Role of Chemotherapy

Multiple Myeloma Detection v. Multiple Myeloma is cancer of the plasma cell ØPlasma cells normally produce antibodies ØOut of control plasma cells produce tumors ØWhen tumors appear in multiple sites they are called Multiple Myeloma v. Dataset Ø 105 patients: 74 with MM, 31 healthy ØEach patient is represented by 7008 gene measurements taken from plasma cell samples ØFor each one of the 7008 gene measurements ØAbsolute Call (AC): ØAbsent (A), Marginal (M) or Present (P) ØAverage Difference (AD): ØPositive or negative number

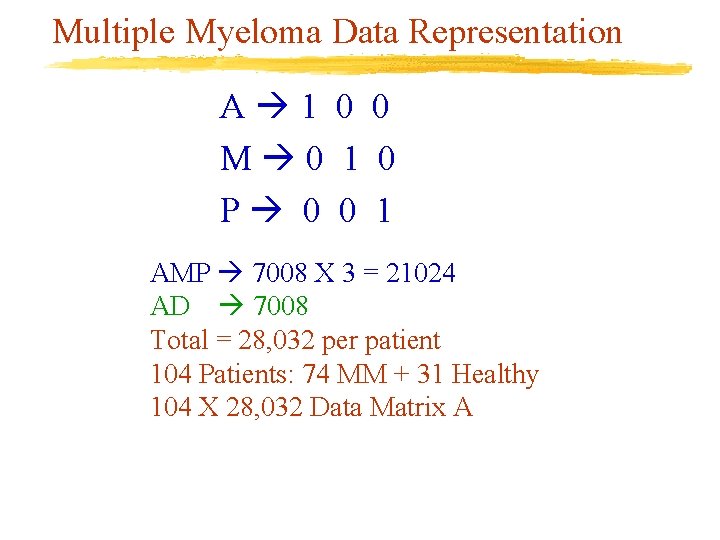
Multiple Myeloma Data Representation A 1 0 0 M 0 1 0 P 0 0 1 AMP 7008 X 3 = 21024 AD 7008 Total = 28, 032 per patient 104 Patients: 74 MM + 31 Healthy 104 X 28, 032 Data Matrix A

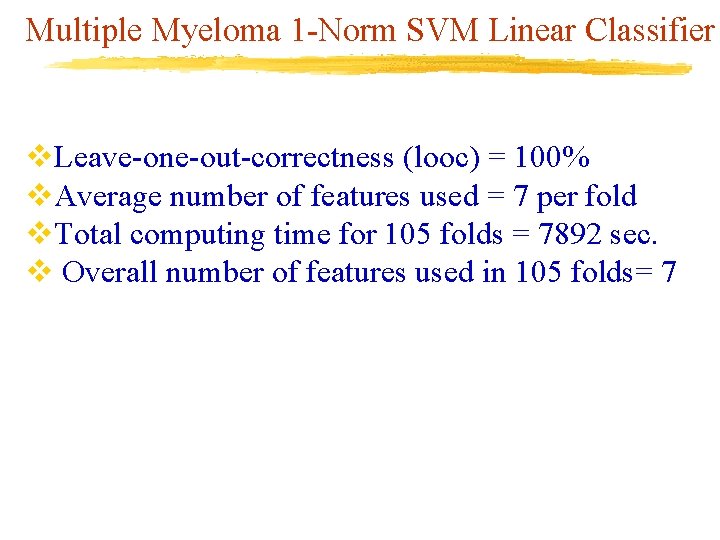
Multiple Myeloma 1 -Norm SVM Linear Classifier v. Leave-one-out-correctness (looc) = 100% v. Average number of features used = 7 per fold v. Total computing time for 105 folds = 7892 sec. v Overall number of features used in 105 folds= 7

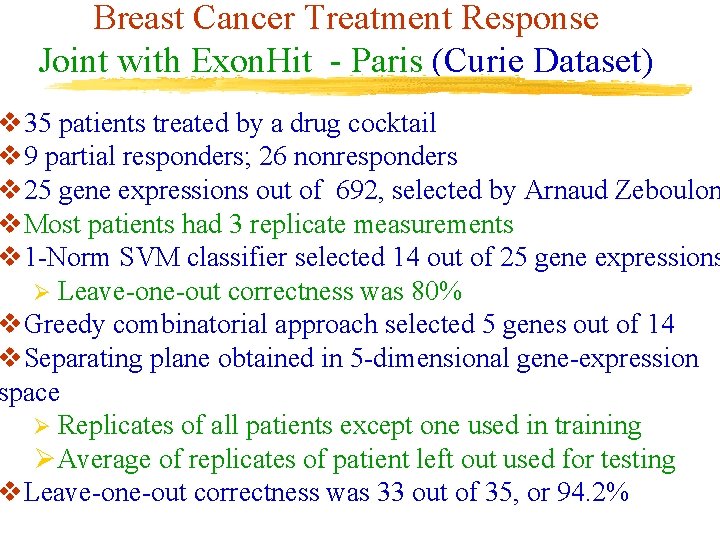
Breast Cancer Treatment Response Joint with Exon. Hit - Paris (Curie Dataset) v 35 patients treated by a drug cocktail v 9 partial responders; 26 nonresponders v 25 gene expressions out of 692, selected by Arnaud Zeboulon v. Most patients had 3 replicate measurements v 1 -Norm SVM classifier selected 14 out of 25 gene expressions Ø Leave-one-out correctness was 80% v. Greedy combinatorial approach selected 5 genes out of 14 v. Separating plane obtained in 5 -dimensional gene-expression space Ø Replicates of all patients except one used in training ØAverage of replicates of patient left out used for testing v. Leave-one-out correctness was 33 out of 35, or 94. 2%

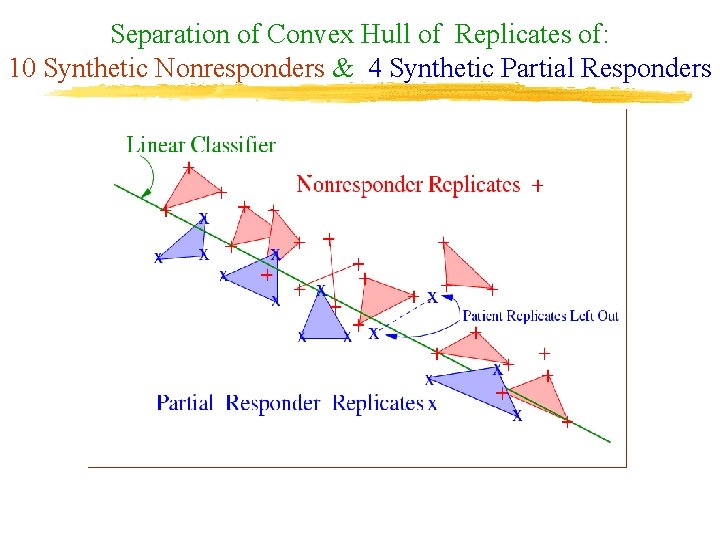
Separation of Convex Hull of Replicates of: 10 Synthetic Nonresponders & 4 Synthetic Partial Responders

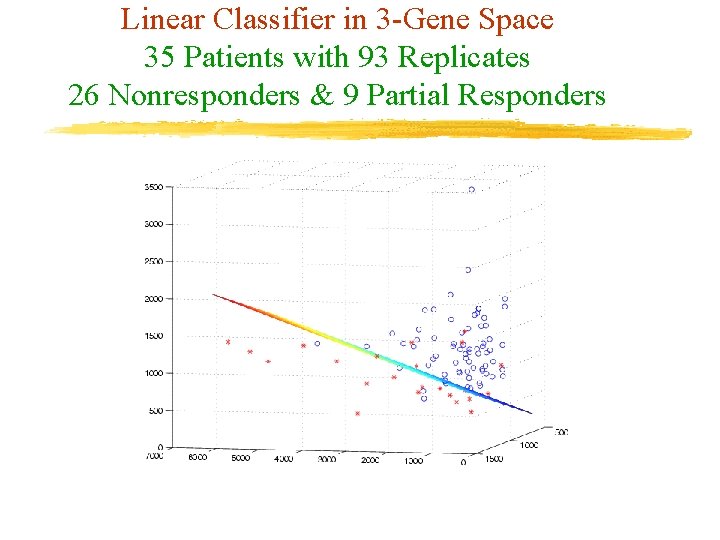
Linear Classifier in 3 -Gene Space 35 Patients with 93 Replicates 26 Nonresponders & 9 Partial Responders

Conclusion v. New approaches for SVM-based classification v. Algorithms capable of classifying data with few examples in very large dimensional spaces v. Typical of microarray classification problems v. Classifiers based on both abstract prior knowledge as well as conventional datasets v. Identification of breast cancer patients that can benefit from chemotherapy v Useful tool for drug discovery