Statistical Methods for Quantitative Trait Loci QTL Mapping



![Interval mapping [Lander and Botstein, 1989] n Consider any one position in the genome Interval mapping [Lander and Botstein, 1989] n Consider any one position in the genome](https://slidetodoc.com/presentation_image/d9d6abb83a1dd6d54c9a99db8a25825e/image-7.jpg)






- Slides: 33

Statistical Methods for Quantitative Trait Loci (QTL) Mapping II Lectures 5 – Oct 12, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12: 00 -1: 20 Johnson Hall (JHN) 022 1

Course Announcements n n HW #1 is out Project proposal n n Due next Wed 1 paragraph describing what you’d like to work on for the class project. 2

Why are we so different? n Any observable characteristic or trait Human genetic diversity n Different “phenotype” n n n TGATCGAAGCTAAATGCATCAGCTGATGATCCTAGC… n Different “genotype” n TGATCGTAGCTAAATGCATCAGCTGATGATCGTAGC… TGATCGCAGCTAAATGCAGCAGCTGATGATCGTAGC… Appearance Disease susceptibility Drug responses : n Individual-specific DNA 3 billion-long string ……ACTGTTAGGCTGAGCTAGCCCAAAATTTATAGC GTCGACTGCAGGGTCCACCAAAGCTCGACTGCAGTCGACGACCTA AAATTTAACCGACTACGAGATGGGCACGTCACTTTTACGCAGCTT GATGATGCTAGCTGATCGTAGCTAAATGCATCAGCTGATGATCGTAGCTAAATGCATCAGCTGA TGATCGTAGCTAAATGCATCAGCTGATTCACTTTTACGCAGCTTG ATGACGACTACGAGATGGGCACGTTCACCATCTACTACTACTCATCAACCAAAAACACTACTCATCATCTACATCTA TCATCATCACATCTACTGGGGGTGGGATAGTGTGCTCGATC 3 GATCGTCAGCTGATCGACGGCAG……

Appearance, Personality, Disease susceptibility, Drug responses, … Motivation n Which sequence variation affects a trait? n n Better understanding disease mechanisms Personalized medicine Sequence variations Instruction Different instruction … AG GTC XXX ACTTCGGAACATATCAAATCCAACGC XX … DNA – 3 billion long! cell A different A person Obese? Bold? Diabetes? Parkinson’s disease? Heart disease? Colon cancer? : 15% 30% 6. 2% 0. 3% 20. 1% 6. 5% 4

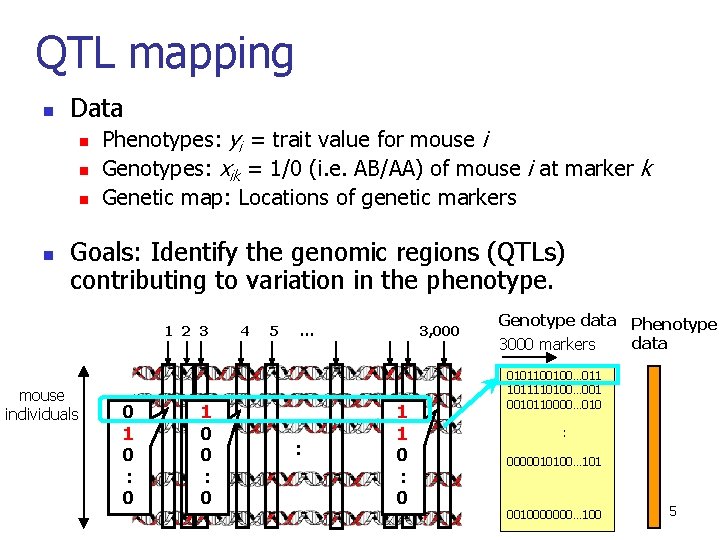
QTL mapping n Data n n Phenotypes: yi = trait value for mouse i Genotypes: xik = 1/0 (i. e. AB/AA) of mouse i at marker k Genetic map: Locations of genetic markers Goals: Identify the genomic regions (QTLs) contributing to variation in the phenotype. 1 2 3 mouse individuals 0 1 0 : 0 1 0 0 : 0 4 5 … : 3, 000 1 1 0 : 0 Genotype data Phenotype data 3000 markers 0101100100… 011 1011110100… 0010110000… 010 : 0000010100… 101 0010000000… 100 5

Outline n Statistical methods for mapping QTL n n What is QTL? Experimental animals Analysis of variance (marker regression) Interval mapping (EM) QTL? 1 2 3 mouse individuals 0 1 0 : 0 1 0 0 : 0 4 5 … : 3, 000 1 1 0 : 0 6
![Interval mapping Lander and Botstein 1989 n Consider any one position in the genome Interval mapping [Lander and Botstein, 1989] n Consider any one position in the genome](https://slidetodoc.com/presentation_image/d9d6abb83a1dd6d54c9a99db8a25825e/image-7.jpg)
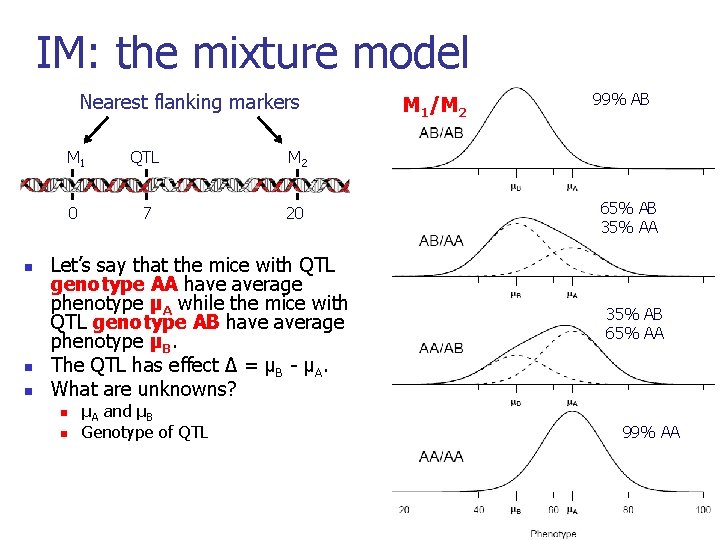
Interval mapping [Lander and Botstein, 1989] n Consider any one position in the genome as the location for a putative QTL. n For a particular mouse, let z = 1/0 if (unobserved) genotype at QTL is AB/AA. n Calculate P(z = 1 | marker data). n n Need only consider nearby genotyped markers. May allow for the presence of genotypic errors. n Given genotype at the QTL, phenotype is distributed as N(µ+∆z, σ2). n Given marker data, phenotype follows a mixture of normal distributions. 7

IM: the mixture model Nearest flanking markers n n n M 1 QTL M 2 0 7 20 Let’s say that the mice with QTL genotype AA have average phenotype µA while the mice with QTL genotype AB have average phenotype µB. The QTL has effect ∆ = µB - µA. What are unknowns? n n µA and µB Genotype of QTL M 1/M 2 99% AB 65% AB 35% AA 35% AB 65% AA 99% AA 8

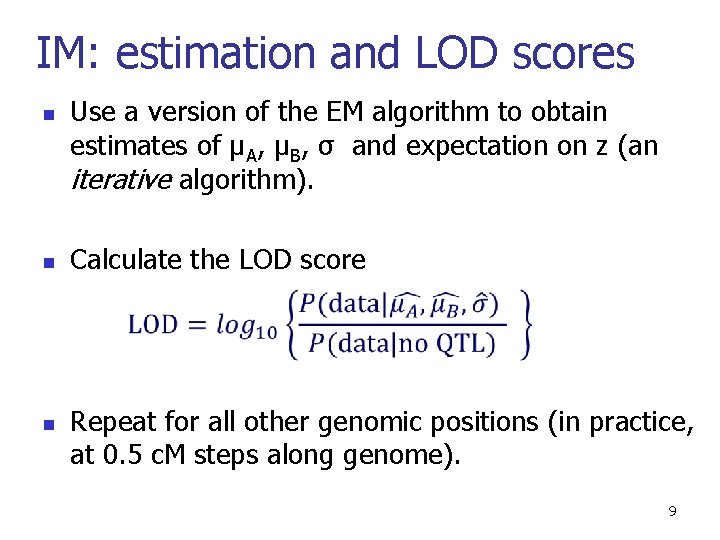
IM: estimation and LOD scores n n Use a version of the EM algorithm to obtain estimates of µA, µB, σ and expectation on z (an iterative algorithm). Calculate the LOD score n Repeat for all other genomic positions (in practice, at 0. 5 c. M steps along genome). 9

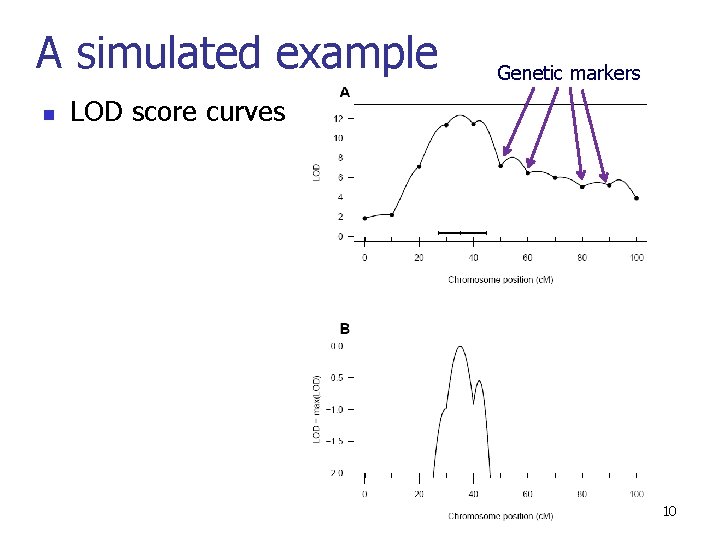
A simulated example n Genetic markers LOD score curves 10

Interval mapping n Advantages n n n Make proper account of missing data Can allow for the presence of genotypic errors Pretty pictures High power in low-density scans Improved estimate of QTL location Disadvantages n n Greater computational effort (doing EM for each position) Requires specialized software More difficult to include covariates Only considers one QTL at a time 11

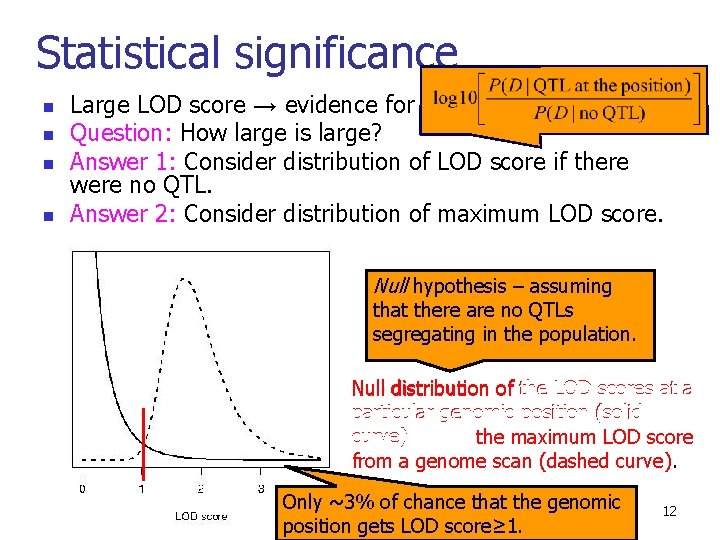
Statistical significance n n Large LOD score → evidence for QTL Question: How large is large? Answer 1: Consider distribution of LOD score if there were no QTL. Answer 2: Consider distribution of maximum LOD score. Null hypothesis – assuming that there are no QTLs segregating in the population. Null distribution of the LOD scores at a particular genomic position (solid curve) and of the maximum LOD score from a genome scan (dashed curve). Only ~3% of chance that the genomic position gets LOD score≥ 1. 12

LOD thresholds n n n To account for the genome-wide search, compare the observed LOD scores to the null distribution of the maximum LOD score, genome-wide, that would be obtained if there were no QTL anywhere. LOD threshold = 95 th percentile of the distribution of genome-wide max LOD, when there are no QTL anywhere. Methods for obtaining thresholds n n n Analytical calculations (assuming dense map of markers) (Lander & Botstein, 1989) Computer simulations Permutation/ randomized test (Churchill & Doerge, 1994) 13

More on LOD thresholds n Appropriate threshold depends on: n n n Size of genome Number of typed markers Pattern of missing data Stringency of significance threshold Type of cross (e. g. F 2 intercross vs backcross) Etc 14

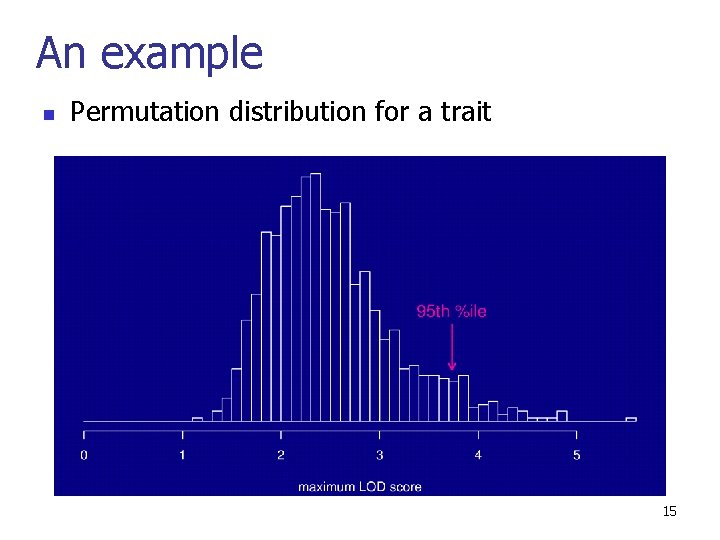
An example n Permutation distribution for a trait 15

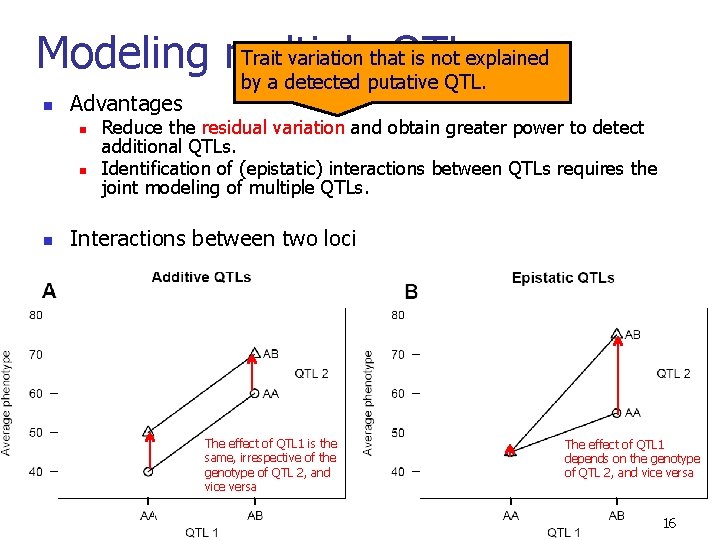
Trait variation that is not explained Modeling multiple QTLs by a detected putative QTL. n Advantages n n n Reduce the residual variation and obtain greater power to detect additional QTLs. Identification of (epistatic) interactions between QTLs requires the joint modeling of multiple QTLs. Interactions between two loci The effect of QTL 1 is the same, irrespective of the genotype of QTL 2, and vice versa The effect of QTL 1 depends on the genotype of QTL 2, and vice versa 16

Multiple marker model n n Let y = phenotype, x = genotype data. Imagine a small number of QTL with genotypes x 1, …, xp n n 2 p or 3 p distinct genotypes for backcross and intercross, respectively We assume that E(y|x) = µ(x 1, …, xp), var(y|x) = σ2(x 1, …, xp) 17

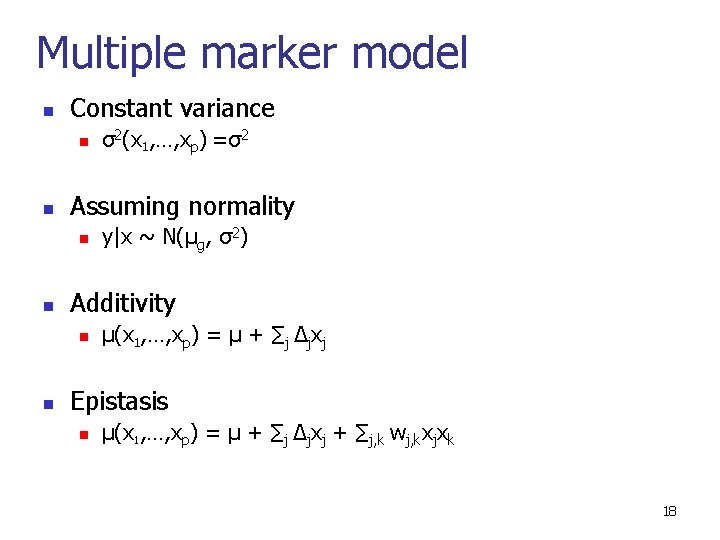
Multiple marker model n Constant variance n n Assuming normality n n y|x ~ N(µg, σ2) Additivity n n σ2(x 1, …, xp) =σ2 µ(x 1, …, xp) = µ + ∑j ∆jxj Epistasis n µ(x 1, …, xp) = µ + ∑j ∆jxj + ∑j, k wj, kxjxk 18

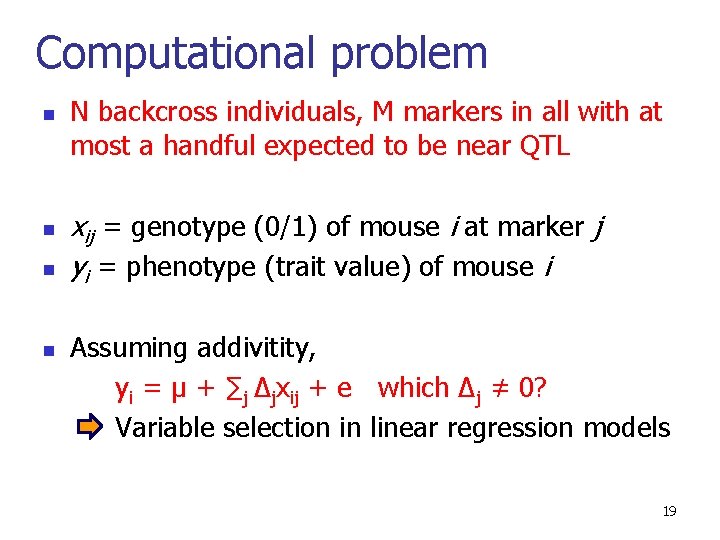
Computational problem n n N backcross individuals, M markers in all with at most a handful expected to be near QTL xij = genotype (0/1) of mouse i at marker j yi = phenotype (trait value) of mouse i Assuming addivitity, yi = µ + ∑j ∆jxij + e which ∆j ≠ 0? Variable selection in linear regression models 19

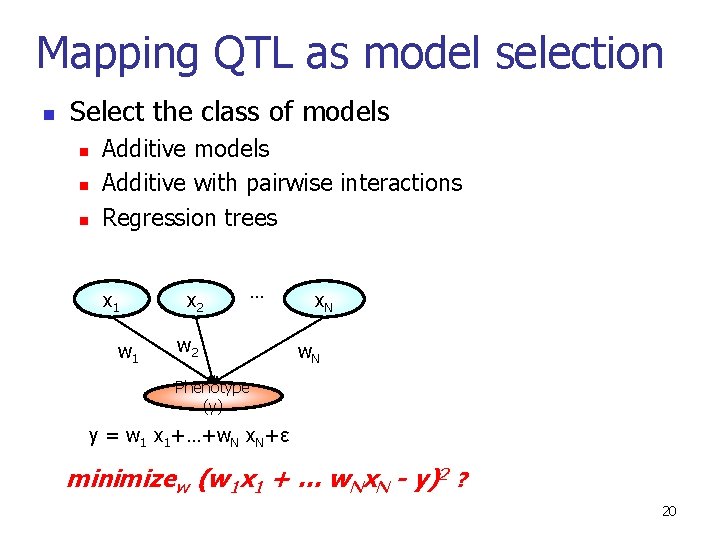
Mapping QTL as model selection n Select the class of models n n n Additive models Additive with pairwise interactions Regression trees x 1 w 1 x 2 … w 2 x. N w. N Phenotype (y) y = w 1 x 1+…+w. N x. N+ε minimizew (w 1 x 1 + … w. Nx. N - y)2 ? 20

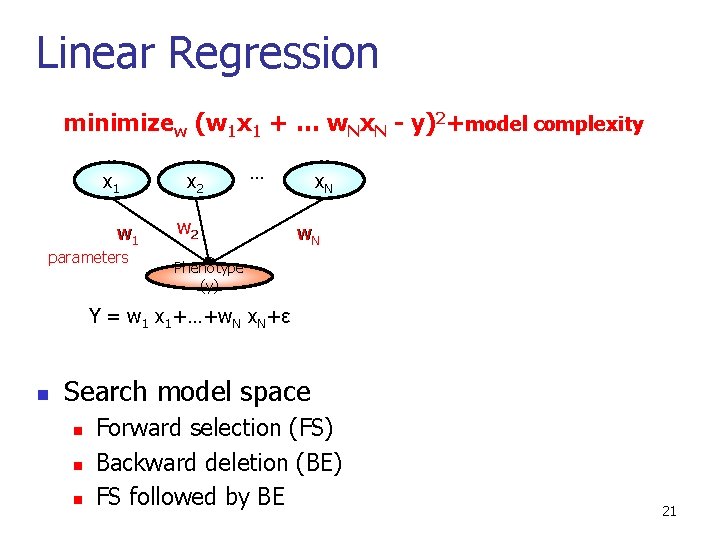
Linear Regression minimizew (w 1 x 1 + … w. Nx. N - y)2+model complexity x 1 w 1 parameters x 2 … w 2 x. N w. N Phenotype (y) Y = w 1 x 1+…+w. N x. N+ε n Search model space n n n Forward selection (FS) Backward deletion (BE) FS followed by BE 21

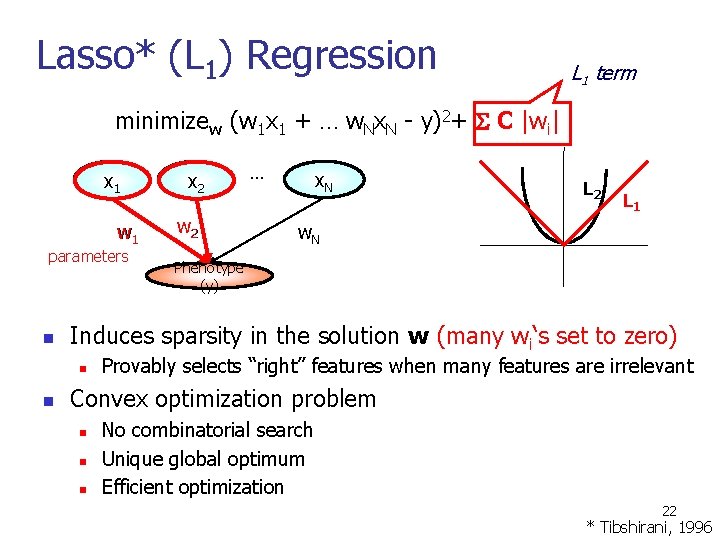
Lasso* (L 1) Regression L 1 term minimizew (w 1 x 1 + … w. Nx. N - y)2+ C |wi| x 1 w 1 parameters n w 2 … x. N L 2 L 1 w. N Phenotype (y) Induces sparsity in the solution w (many wi‘s set to zero) n n x 2 Provably selects “right” features when many features are irrelevant Convex optimization problem n n n No combinatorial search Unique global optimum Efficient optimization 22 * Tibshirani, 1996

Model selection n Compare models n n Likelihood function + model complexity (eg # QTLs) Cross validation test Sequential permutation tests Assess performance n n Maximize the number of QTL found Control the false positive rate 23

Outline n Basic concepts n n Haplotype, haplotype frequency Recombination rate Linkage disequilibrium Haplotype reconstruction n n Parsimony-based approach EM-based approach 24

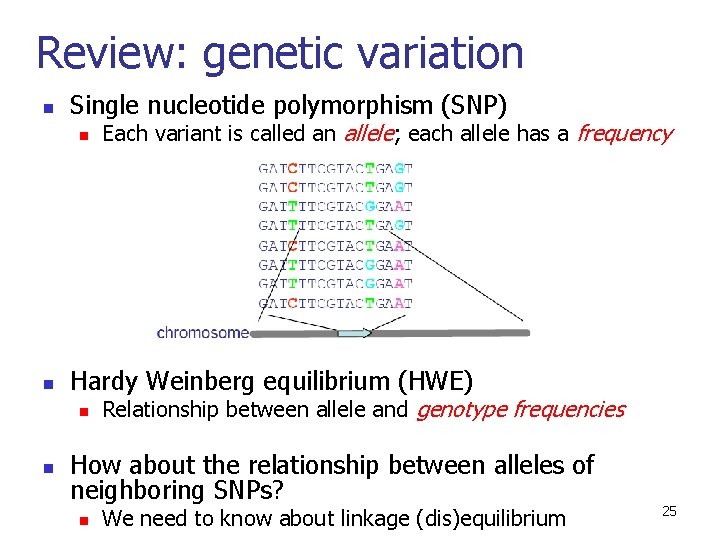
Review: genetic variation n Single nucleotide polymorphism (SNP) n n Hardy Weinberg equilibrium (HWE) n n Each variant is called an allele; each allele has a frequency Relationship between allele and genotype frequencies How about the relationship between alleles of neighboring SNPs? n We need to know about linkage (dis)equilibrium 25

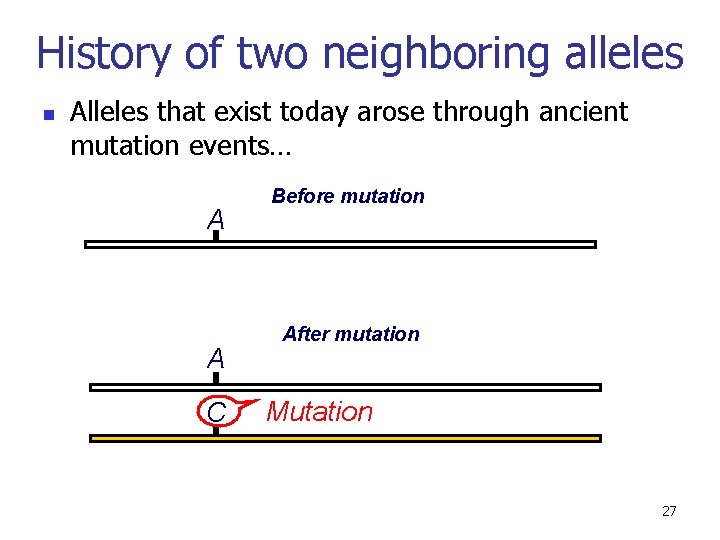
Let’s consider the history of two neighboring alleles… 26

History of two neighboring alleles n Alleles that exist today arose through ancient mutation events… A A C Before mutation After mutation Mutation 27

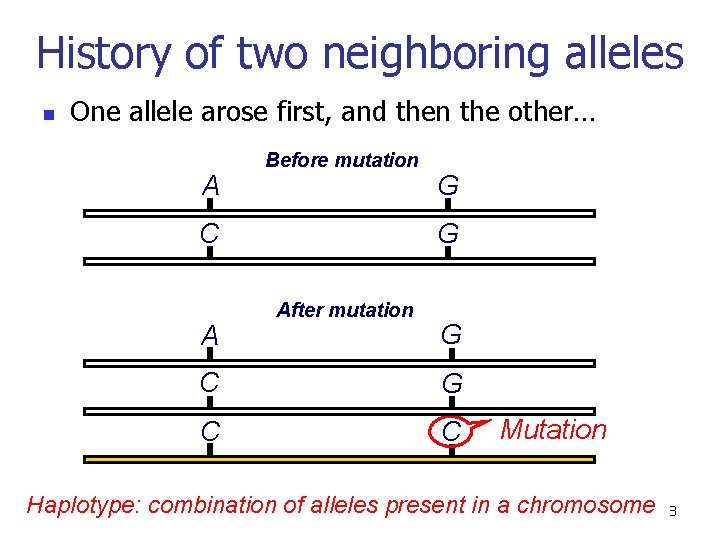
History of two neighboring alleles n One allele arose first, and then the other… A Before mutation C A G G After mutation G C C Mutation Haplotype: combination of alleles present in a chromosome 28

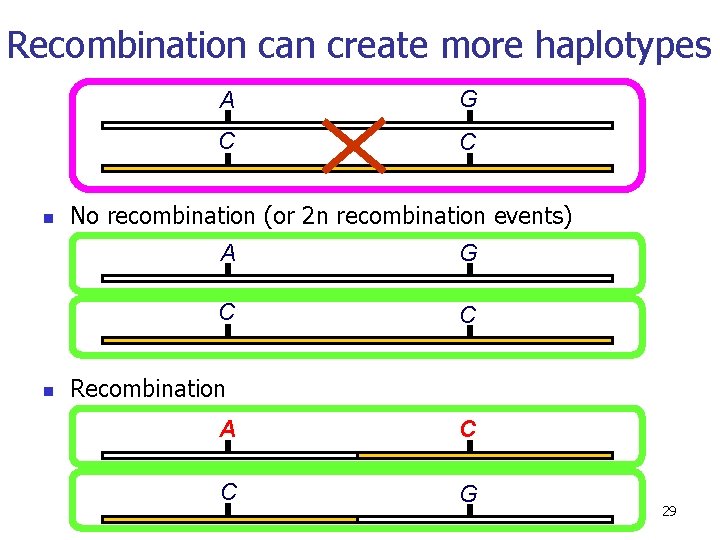
Recombination can create more haplotypes n n A G C C No recombination (or 2 n recombination events) A G C C Recombination A C C G 29

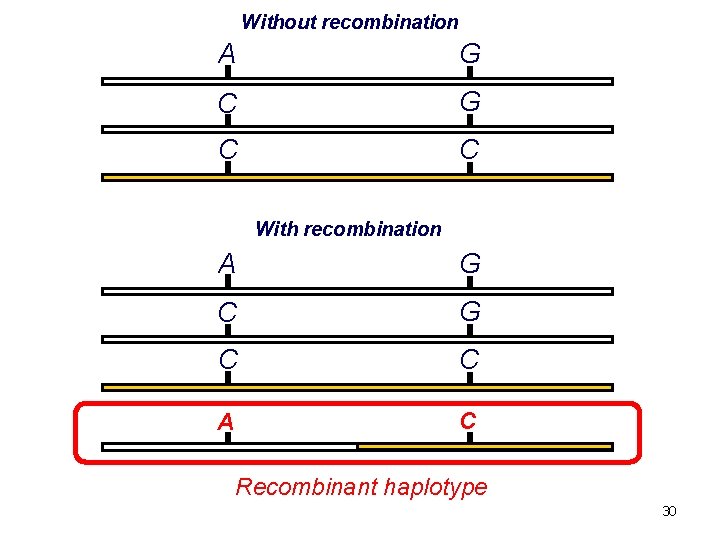
Without recombination A G C C With recombination A G C C A C Recombinant haplotype 30

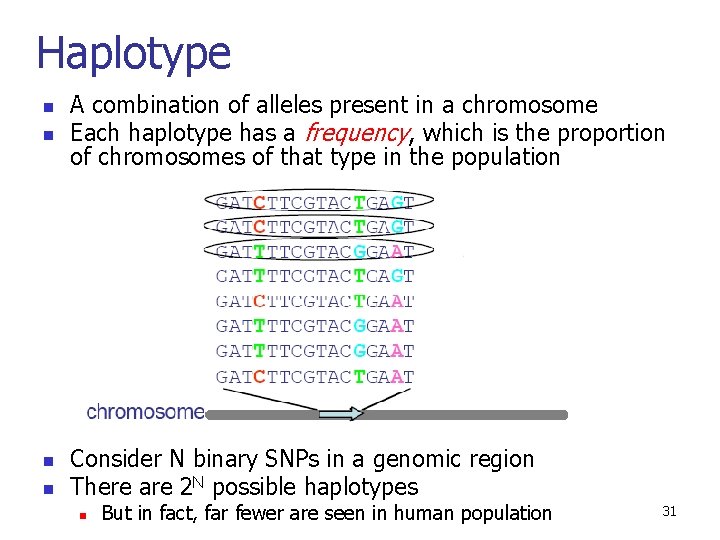
Haplotype n n A combination of alleles present in a chromosome Each haplotype has a frequency, which is the proportion of chromosomes of that type in the population Consider N binary SNPs in a genomic region There are 2 N possible haplotypes n But in fact, far fewer are seen in human population 31

More on haplotype n What determines haplotype frequencies? n n Linkage disequilibrium (LD) n n Recombination rate (r) between neighboring alleles Depends on the population r is different for different regions in genome Non-random association of alleles at two or more loci, not necessarily on the same chromosome. Why do we care about haplotypes or LD? 32

References n n n Prof Goncalo Abecasis (Univ of Michigan)’s lecture note Broman, K. W. , Review of statistical methods for QTL mapping in experimental crosses Doerge, R. W. , et al. Statistical issues in the search for genes affecting quantitative traits in experimental populations. Stat. Sci. ; 12: 195 -219, 1997. Lynch, M. and Walsh, B. Genetics and analysis of quantitative traits. Sinauer Associates, Sunderland, MA, pp. 431 -89, 1998. Broman, K. W. , Speed, T. P. A review of methods for identifying QTLs in experimental crosses, 1999. 33