Trait Mapping Recombination Mapping SNP mapping BIO 520

Trait Mapping • Recombination Mapping • SNP mapping BIO 520 Bioinformatics Jim Lund
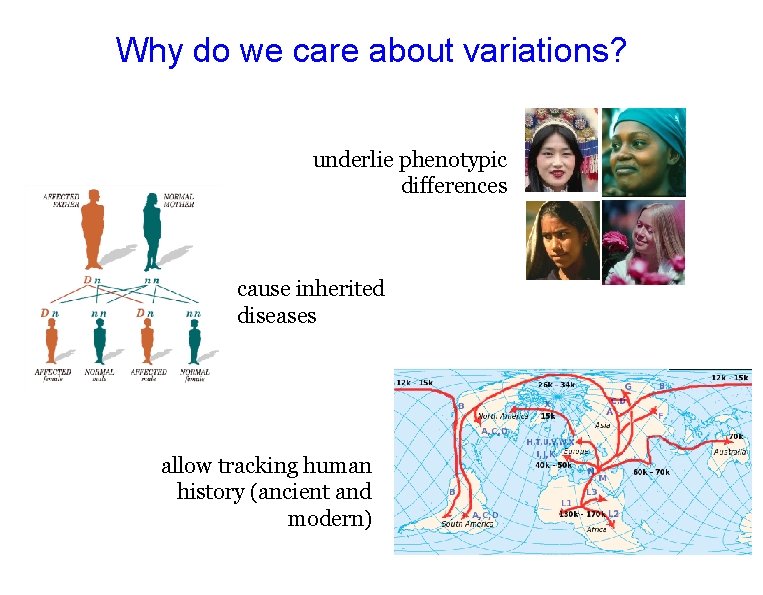
Why do we care about variations? underlie phenotypic differences cause inherited diseases allow tracking human history (ancient and modern)
Traits • Mendelian – single locus, few alleles – high penetrance, high expressivity – eg color, enzyme, molecular, genetic diseases (CF, hemophilia…) • Quantitative – – multiple allele, multilocus variable penetrance, expressivity epistasis, environmental effects eg. blood pressure, weight, IQ. . .

Traits How do we find their basis? • Association of variance in trait with variance in gene • Genetic linkage
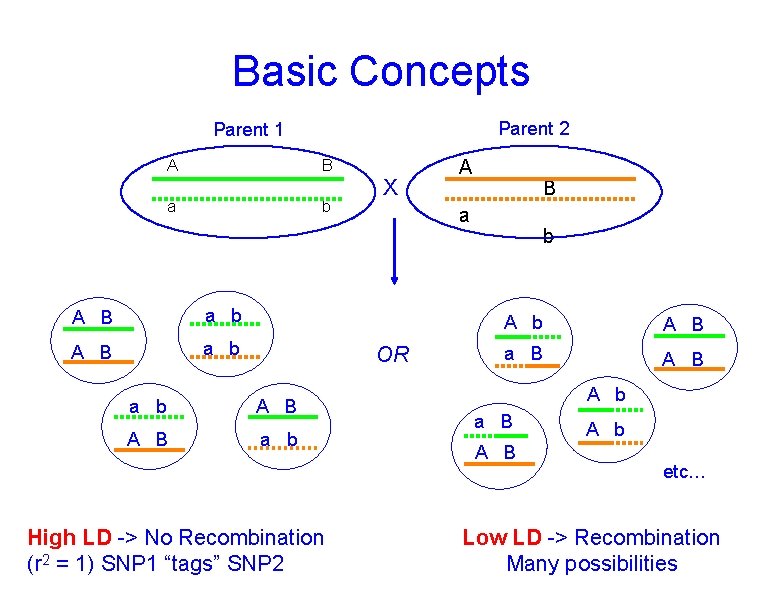
Basic Concepts Parent 2 Parent 1 A B a b A B a b X A B a b High LD -> No Recombination (r 2 = 1) SNP 1 “tags” SNP 2 B a OR a b A B a B A b etc… Low LD -> Recombination Many possibilities

Mapping Issues • Need many arbitrary, polymorphic markers for dense map – Molecular markers: RFLP, STS, SNP • Need many progeny – 100 progeny for 1 c. M map – 1000/0. 1 c. M map, 100 kb in mouse • Map distance varies (the ratio of kb/c. M not constant) – centromere suppression – inversion suppression
Genetic crosses • Model organisms, e. g. Fungi, no problem • Humans – rare woman who will bear >5, >10 children – controlled breeding problematic

Alternate Mapping • Pedigree analyses – likelihood estimation – The original method, now less common • Population-based mapping – association studies – linkage disequilibrium
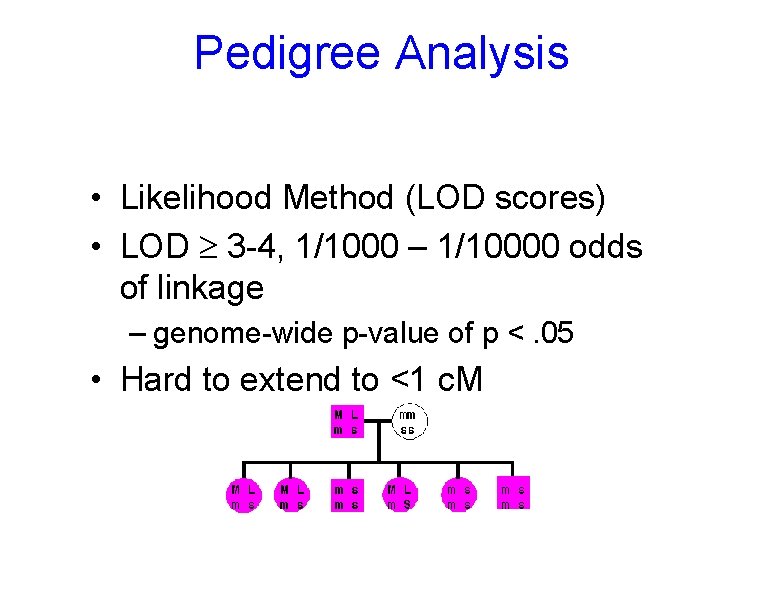
Pedigree Analysis • Likelihood Method (LOD scores) • LOD 3 -4, 1/1000 – 1/10000 odds of linkage – genome-wide p-value of p <. 05 • Hard to extend to <1 c. M
Cloning Human Genes • • Positional/Candidate Only Functional
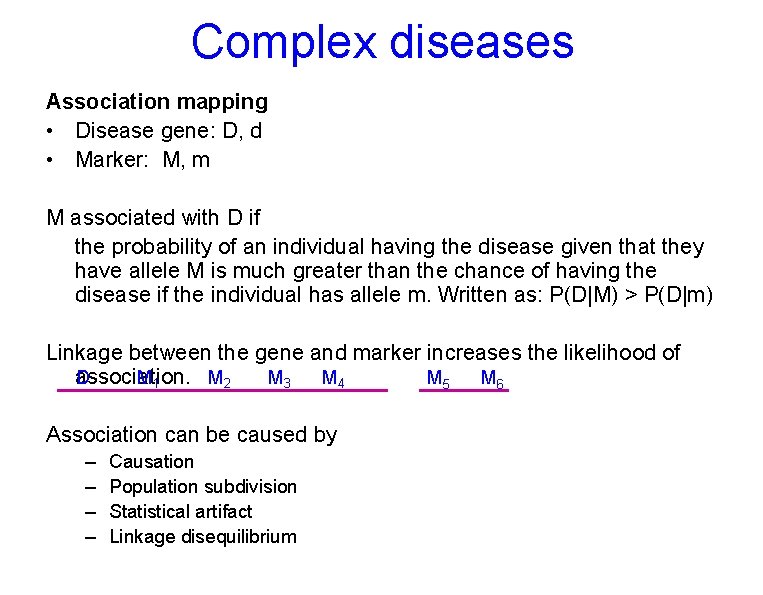
Complex diseases Association mapping • Disease gene: D, d • Marker: M, m M associated with D if the probability of an individual having the disease given that they have allele M is much greater than the chance of having the disease if the individual has allele m. Written as: P(D|M) > P(D|m) Linkage between the gene and marker increases the likelihood of D M 1 M 2 M 3 M 4 association. M 5 M 6 Association can be caused by – – Causation Population subdivision Statistical artifact Linkage disequilibrium
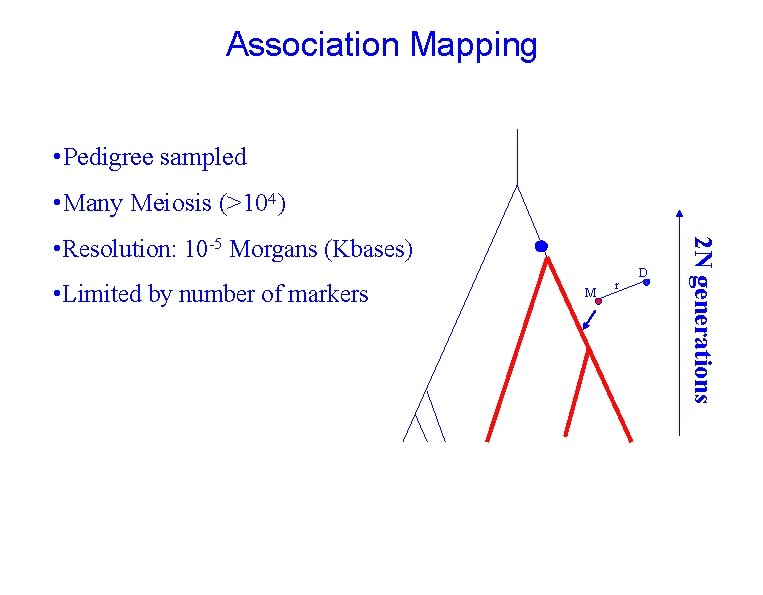
Association Mapping • Pedigree sampled • Many Meiosis (>104) • Limited by number of markers M r D 2 N generations • Resolution: 10 -5 Morgans (Kbases)
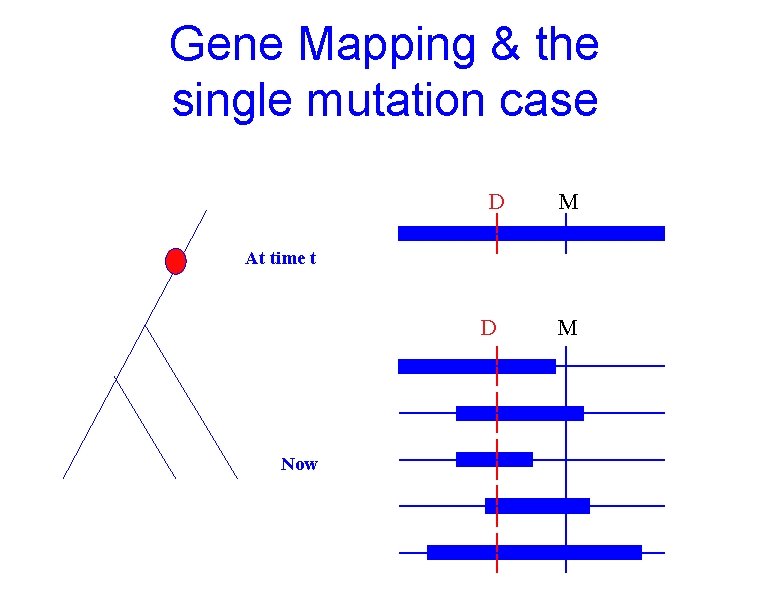
Gene Mapping & the single mutation case D M At time t D Now M
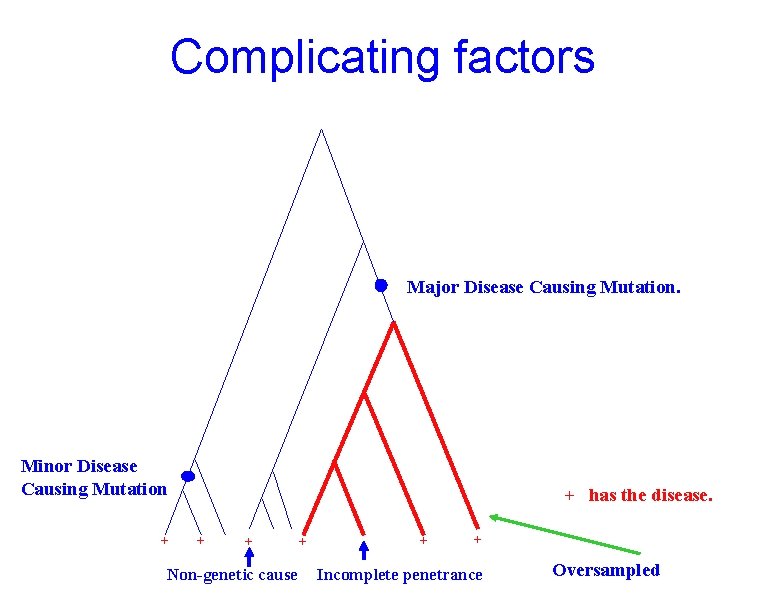
Complicating factors Major Disease Causing Mutation. Minor Disease Causing Mutation + + has the disease. + + Non-genetic cause + + + Incomplete penetrance Oversampled
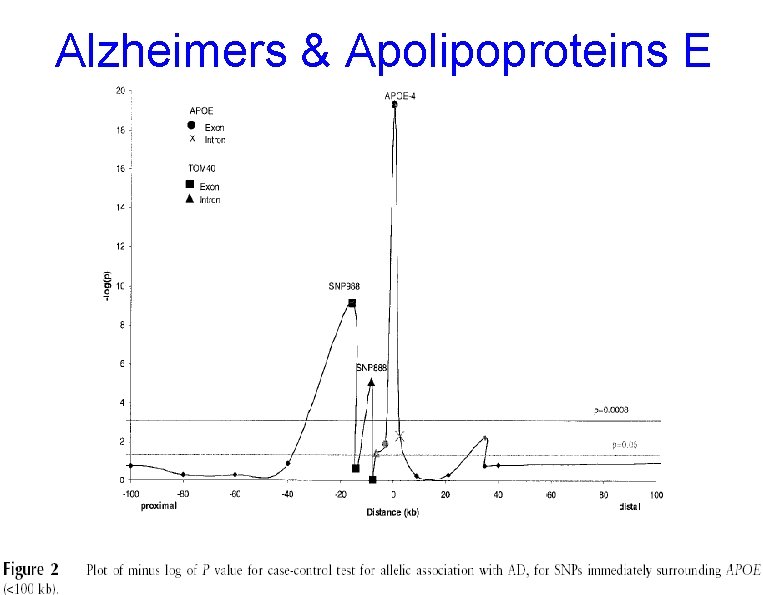
Alzheimers & Apolipoproteins E

Definition of QTL? A quantitative trait locus (QTL) is the location of individual or multiple loci that affects a trait that is measured on a quantitative (linear) scale. Examples of quantitative traits are blood pressure and grain yield (measured on a balance). These traits are typically affected by more than one gene, and also by the environment. Thus, mapping QTL is not as simple as mapping a single gene that affects a qualitative trait (such as an inborn error of metabolism). http: //gnome. agrenv. mcgill. ca/tinker/pgiv/whatis. htm

QTLs-interesting traits • Heritability often ~0. 5 • Traits like: – Heart disease – Depression – Type II diabetes – High blood pressure – Arthritis – Most diseases!

QTLs-simple problems • 30, 000 markers – P-value=0. 01 – 299 false hits, 1 real one – Correct for multiple testing • 2 QTLS near one another – “ghost” QTL between them

Factors that lead to success in mapping QTLs • Simple, easily quantified trait • Genes of major effect – distinct chromosomal loci • Well-defined map • Large numbers of progeny – inbred – outbred
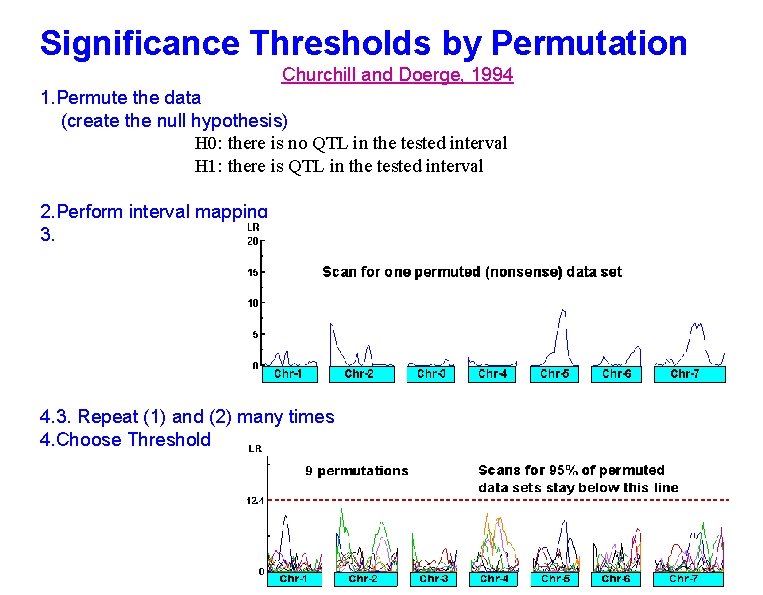
Significance Thresholds by Permutation Churchill and Doerge, 1994 1. Permute the data (create the null hypothesis) H 0: there is no QTL in the tested interval H 1: there is QTL in the tested interval 2. Perform interval mapping 3. 4. 3. Repeat (1) and (2) many times 4. Choose Threshold

Human SNPs • About 10 million SNPs exist in human populations where the rarer SNP allele has a frequency of at least 1%. • A set of associated SNP alleles in a region of a chromosome is called a "haplotype". • SNPs are arranged in groups – SNPs within groups show little recombination – Nonrandom association of SNPs results in only a few common haplotypes – Patterns capture most of the variation in a region • The Hap. Map will describe the common patterns of genetic variation in humans. • The Hap. Map Project will identify the associations between SNPs and identify the SNPs that tag them (tag. SNPs).
SNPs identification methods • Pairwise sequence comparison • Deep resequencing • High throughput mismatch detection methods – Denaturing high-performance liquid chromatography (DHPLC) – Single-strand Conformational Polymorphism (SSCP)
Hap. Map • Blocks of adjacent SNPs that show little recombination are called haplotype blocks. • Mean haplotype block length is tens of kb. • Hap. Map project started examining 270 individuals from 4 ethnic groups. • Now expanding to a more comprehensive sample. Characterization of haplotype blocks means that fewer SNPs will need to be typed. 500, 000 SNPs will identify 90% of haplotype blocks.
Hap. Map Glossary • LD (linkage disequilibrium): For a pair of SNP alleles, it’s a measure of deviation from random association (i. e. , a measure of lack of recombination). Measured by D’, r 2, LOD • Phased haplotypes: Estimated distribution of SNP alleles. Alleles transmitted from Mom are in same chromosome haplotype, while Dad’s form the paternal haplotype. • Tag SNPs: Minimum SNP set to identify a haplotype. r 2= 1 indicates two SNPs are redundant, so each one perfectly “tags” the other.
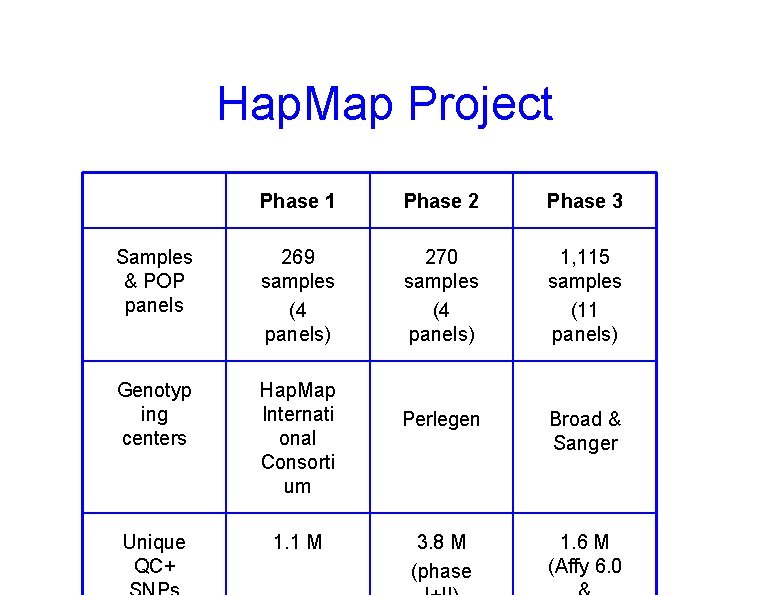
Hap. Map Project Phase 1 Phase 2 Phase 3 Samples & POP panels 269 samples (4 panels) 270 samples (4 panels) 1, 115 samples (11 panels) Genotyp ing centers Hap. Map Internati onal Consorti um Perlegen Broad & Sanger Unique QC+ 1. 1 M 3. 8 M (phase 1. 6 M (Affy 6. 0

Phase 3 Samples * Population is made of family trios

SNP databases • db. SNP (NCBI) – 12 million human SNPs – 5 million validated SNPs – http: //www. ncbi. nlm. nih. gov/SNP/get_html. cgi? which. Html=overview • SNP frequency information • Mapped to the current genome build • Hap. Map (haplotypes)
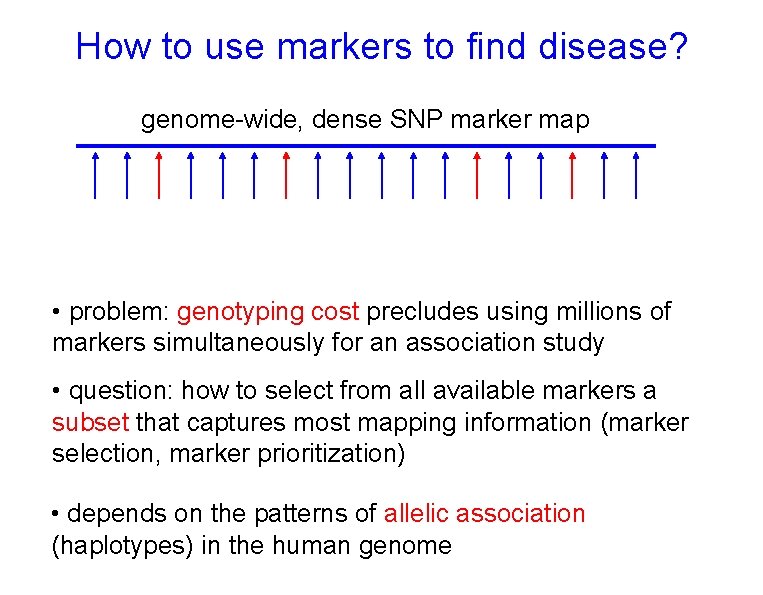
How to use markers to find disease? genome-wide, dense SNP marker map • problem: genotyping cost precludes using millions of markers simultaneously for an association study • question: how to select from all available markers a subset that captures most mapping information (marker selection, marker prioritization) • depends on the patterns of allelic association (haplotypes) in the human genome
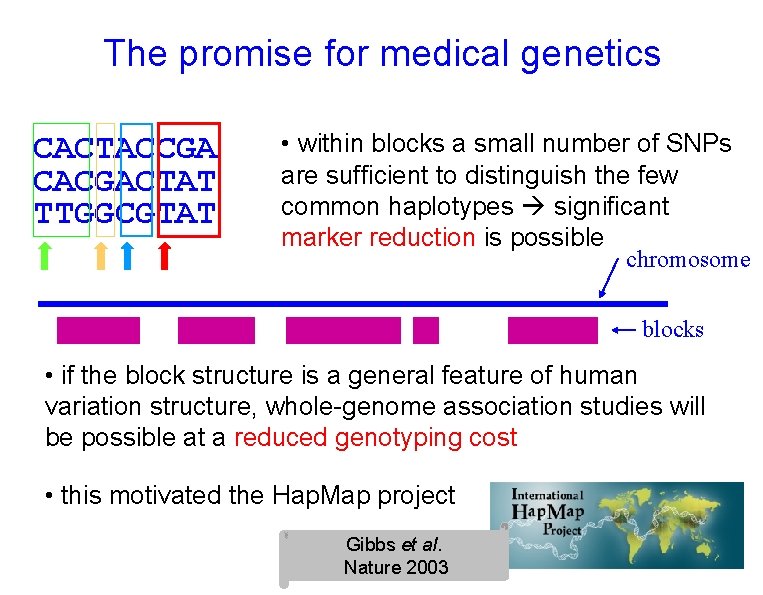
The promise for medical genetics CACTACCGA CACGACTAT TTGGCGTAT • within blocks a small number of SNPs are sufficient to distinguish the few common haplotypes significant marker reduction is possible chromosome blocks • if the block structure is a general feature of human variation structure, whole-genome association studies will be possible at a reduced genotyping cost • this motivated the Hap. Map project Gibbs et al. Nature 2003

The promise for medical genetics • Discover genes contributing to complex diseases • Use these markers to test for inherited disease risk • Find SNPs associated with drug side effects • Make drugs safer. • Rescue drugs abandoned due to significant side effects.

Pathway of Drug Development • Lead or Target (Clinical Candidate) • Animal Model Testing – Toxicity, Efficacy • Phase I Pre-Clinical (toxicity) • Phase II (efficacy) • Phase III (efficacy) • NDA (new drug application) • $100 M 2000 • $0. 5 M 100 • $0. 5 M • $50 M 20 3 2 1

Why pharmacogenomics? • Where do you find the next profitable drug? – The 19/20 drugs that failed AFTER phase 1, but are still efficacious! • How do you decrease the cost of clinical trials? – Don’t enroll people of the “wrong” genotype! • Only give drugs to patients likely to benefit and at a low genetic risk of side effects!
- Slides: 32