Introduction to Quantitative Trait Loci Linkage and Association

















- Slides: 42

Introduction to Quantitative Trait Loci Linkage and Association Studies Lon Cardon Wellcome Trust Centre for Human Genetics University of Oxford Pak Sham Institute of Psychiatry King’s College London Stacey Cherny Both, and then some

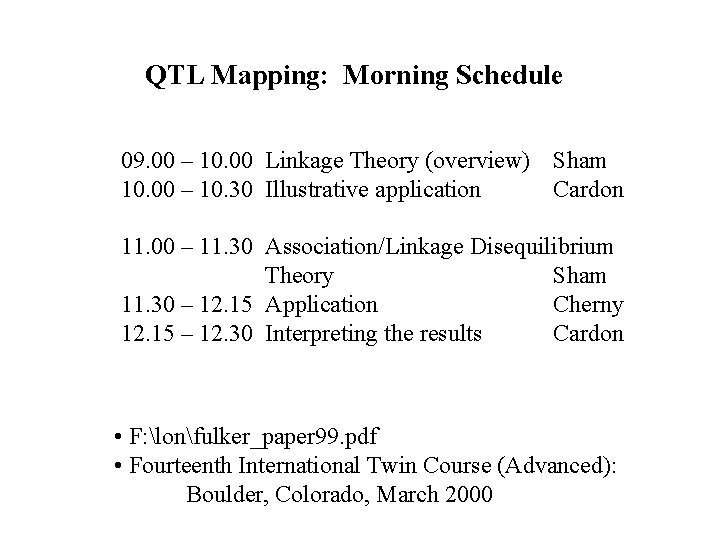
QTL Mapping: Morning Schedule 09. 00 – 10. 00 Linkage Theory (overview) Sham 10. 00 – 10. 30 Illustrative application Cardon 11. 00 – 11. 30 Association/Linkage Disequilibrium Theory Sham 11. 30 – 12. 15 Application Cherny 12. 15 – 12. 30 Interpreting the results Cardon • F: lonfulker_paper 99. pdf • Fourteenth International Twin Course (Advanced): Boulder, Colorado, March 2000

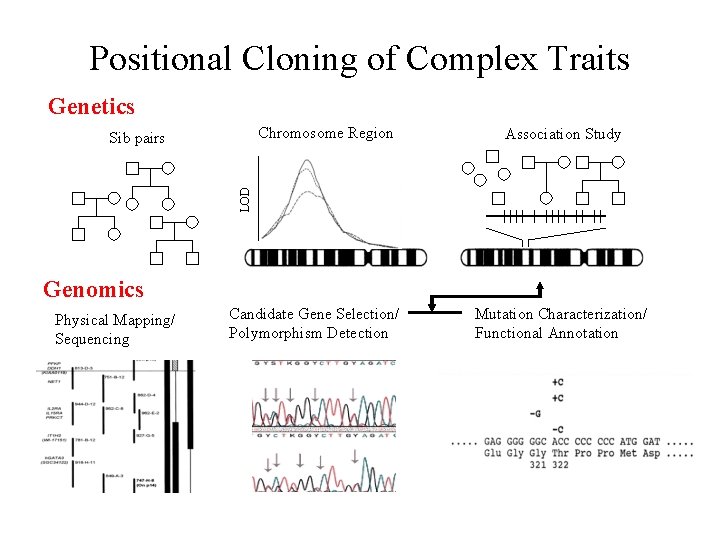
Positional Cloning of Complex Traits Genetics Chromosome Region Association Study LOD Sib pairs Genomics Physical Mapping/ Sequencing Candidate Gene Selection/ Polymorphism Detection Mutation Characterization/ Functional Annotation

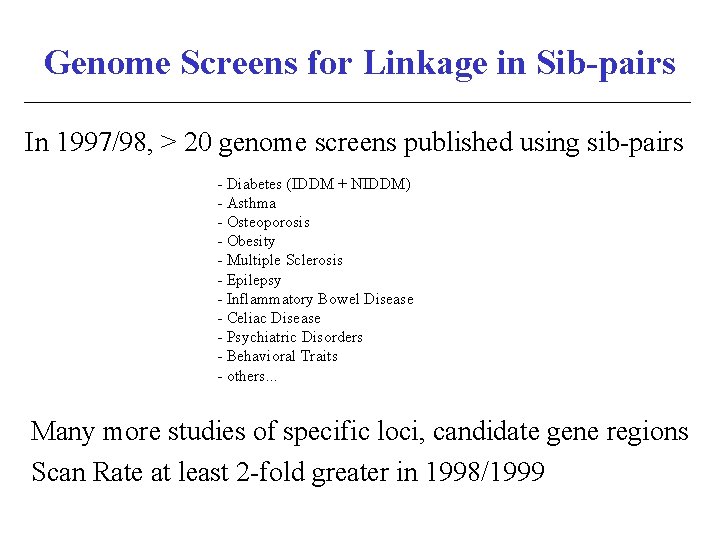
Genome Screens for Linkage in Sib-pairs In 1997/98, > 20 genome screens published using sib-pairs - Diabetes (IDDM + NIDDM) - Asthma - Osteoporosis - Obesity - Multiple Sclerosis - Epilepsy - Inflammatory Bowel Disease - Celiac Disease - Psychiatric Disorders - Behavioral Traits - others. . . Many more studies of specific loci, candidate gene regions Scan Rate at least 2 -fold greater in 1998/1999

Disequilibrium Mapping • 100’s candidate gene studies every year • Replications rare • Genome-wide SNP maps expected in late 2001 (300, 000 SNPs; ~ 1 SNP/10 kb) • Applications in epidemiology, drug design, functional assessment, …

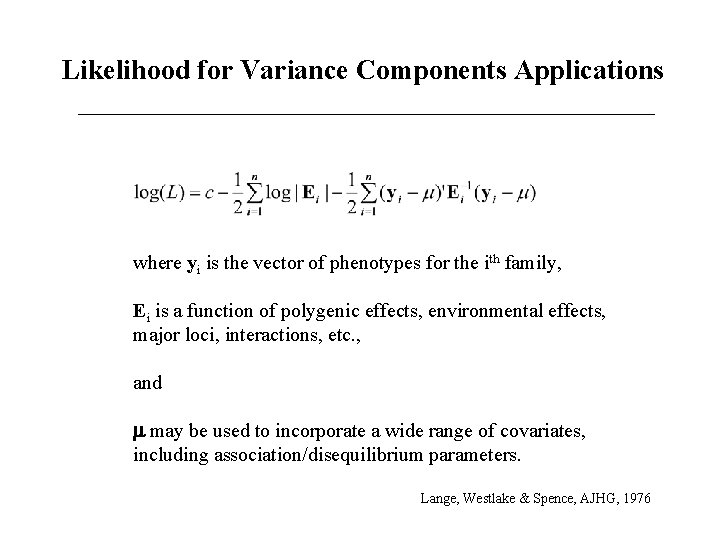
Likelihood for Variance Components Applications where yi is the vector of phenotypes for the ith family, Ei is a function of polygenic effects, environmental effects, major loci, interactions, etc. , and m may be used to incorporate a wide range of covariates, including association/disequilibrium parameters. Lange, Westlake & Spence, AJHG, 1976

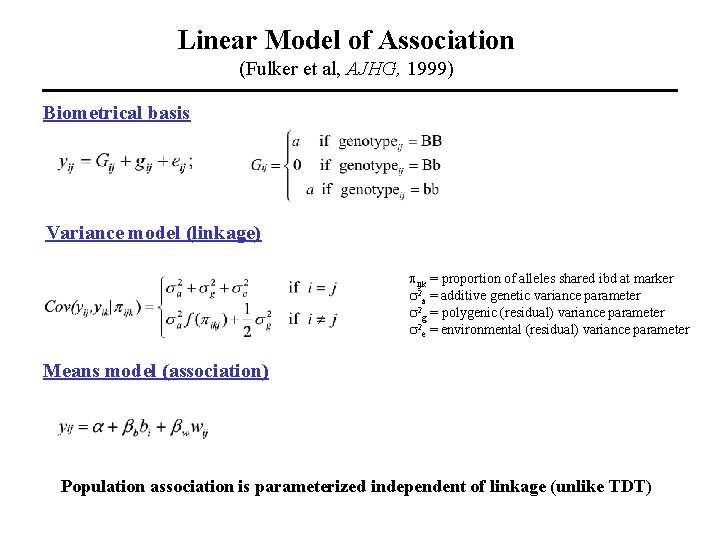
Linear Model of Association (Fulker et al, AJHG, 1999) Biometrical basis Variance model (linkage) pijk = proportion of alleles shared ibd at marker s 2 a = additive genetic variance parameter s 2 g = polygenic (residual) variance parameter s 2 e = environmental (residual) variance parameter Means model (association) Population association is parameterized independent of linkage (unlike TDT)


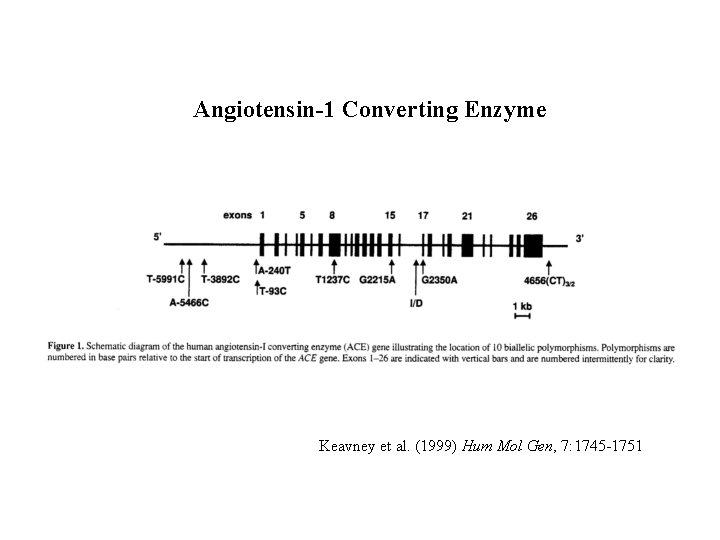
Application: ACE • British population • Circulating ACE levels – Normalized separately for males / females • 10 di-allelic polymorphisms – 26 kb – Common – In strong Linkage disequilibrium • Keavney et al, HMG, 1998

Angiotensin-1 Converting Enzyme Keavney et al. (1999) Hum Mol Gen, 7: 1745 -1751

Angiotensin-1 Converting Enzyme Keavney et al. (1999) Families 83 extended families 4 - 18 members/family age: 19 -90 years Families ascertained for study of blood pressure Phenotype: Plasma ACE activity, standardized within gender No correlation between ACE and SBP or DBP

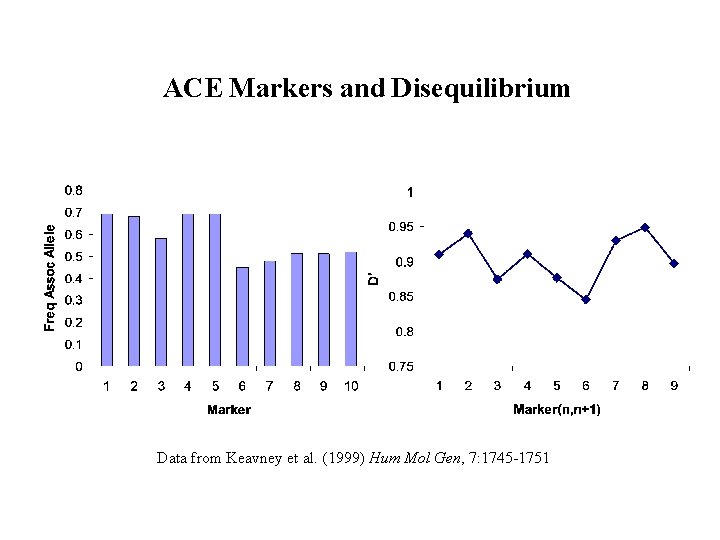
ACE Markers and Disequilibrium Data from Keavney et al. (1999) Hum Mol Gen, 7: 1745 -1751

Angiotensin Converting Enzyme Marker/IBD Files • F: lon2000linkage. mx • F: lon2000marker*. mx



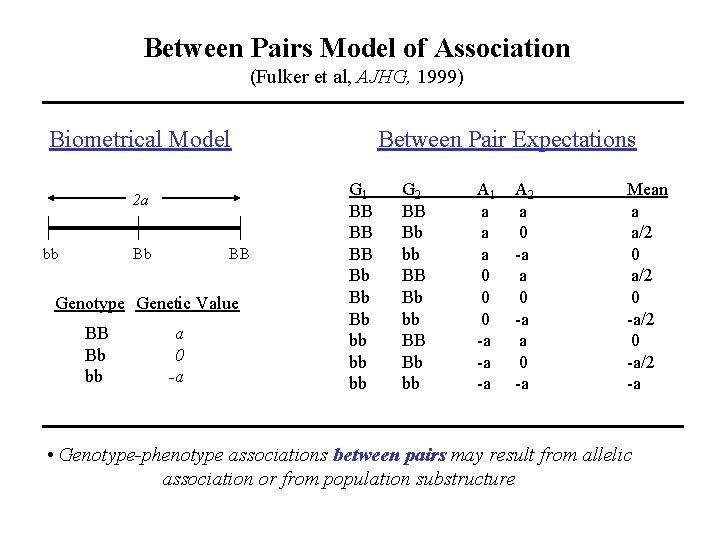
Between Pairs Model of Association (Fulker et al, AJHG, 1999) Biometrical Model 2 a bb Bb BB Genotype Genetic Value BB Bb bb a 0 -a Between Pair Expectations G 1 BB BB BB Bb Bb Bb bb bb bb G 2 BB Bb bb A 1 a a a 0 0 0 -a -a -a A 2 a 0 -a Mean a a/2 0 -a/2 -a • Genotype-phenotype associations between pairs may result from allelic association or from population substructure

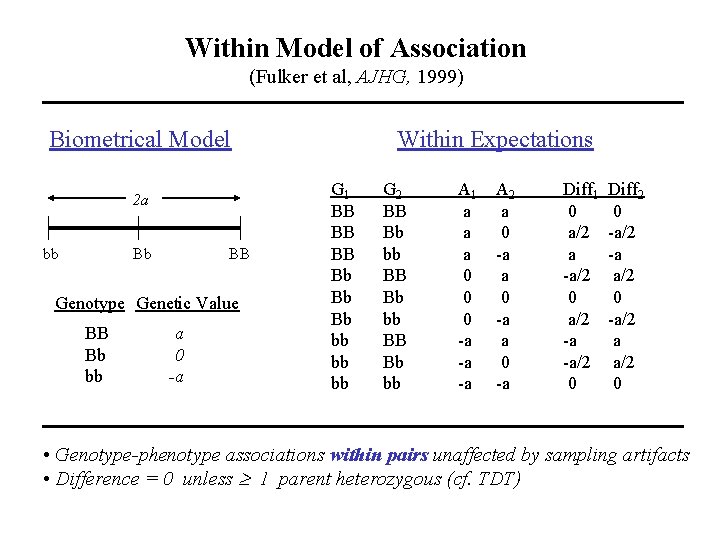
Within Model of Association (Fulker et al, AJHG, 1999) Biometrical Model 2 a bb Bb BB Genotype Genetic Value BB Bb bb a 0 -a Within Expectations G 1 BB BB BB Bb Bb Bb bb bb bb G 2 BB Bb bb A 1 a a a 0 0 0 -a -a -a A 2 a 0 -a Diff 1 0 a/2 a -a/2 0 a/2 -a -a/2 0 Diff 2 0 -a/2 -a a/2 0 -a/2 a a/2 0 • Genotype-phenotype associations within pairs unaffected by sampling artifacts • Difference = 0 unless 1 parent heterozygous (cf. TDT)

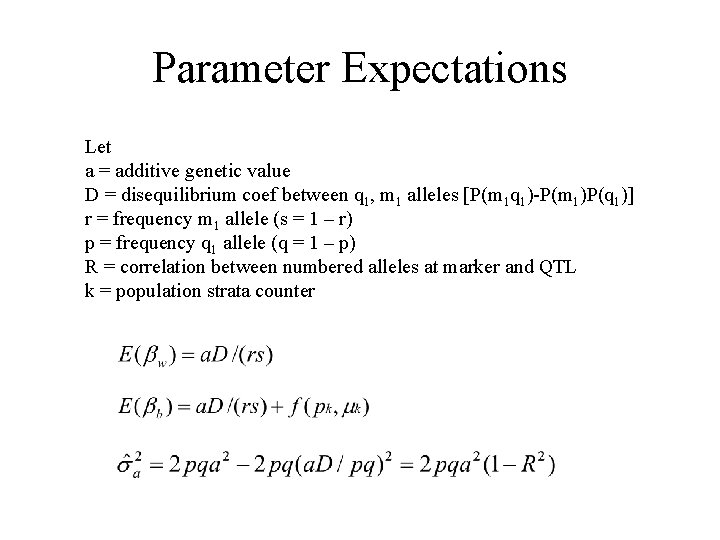
Parameter Expectations Let a = additive genetic value D = disequilibrium coef between q 1, m 1 alleles [P(m 1 q 1)-P(m 1)P(q 1)] r = frequency m 1 allele (s = 1 – r) p = frequency q 1 allele (q = 1 – p) R = correlation between numbered alleles at marker and QTL k = population strata counter

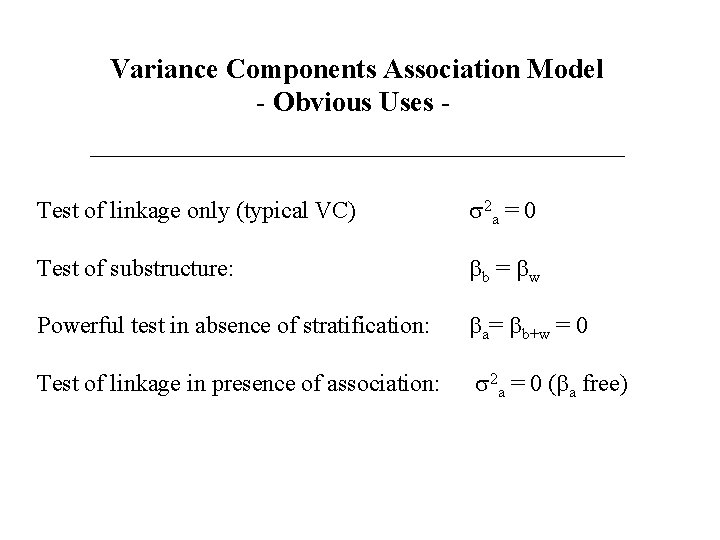
Variance Components Association Model - Obvious Uses - Test of linkage only (typical VC) s 2 a = 0 Test of substructure: bb = bw Powerful test in absence of stratification: ba= bb+w = 0 Test of linkage in presence of association: s 2 a = 0 (ba free)


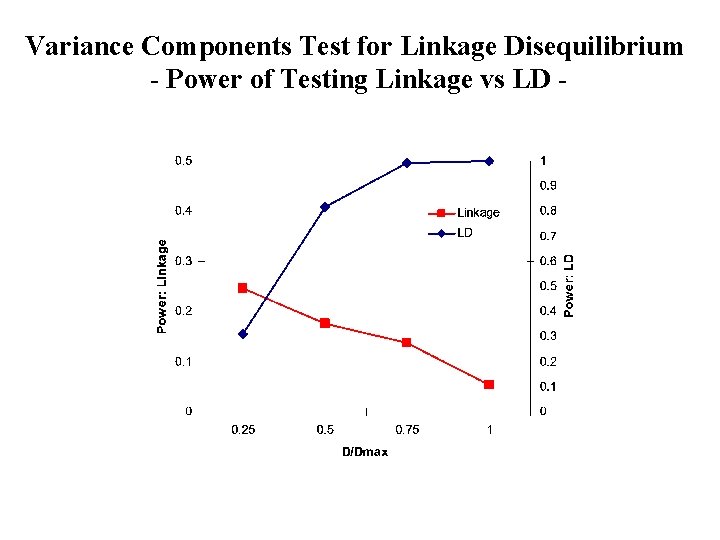
Variance Components Test for Linkage Disequilibrium - Power of Testing Linkage vs LD -





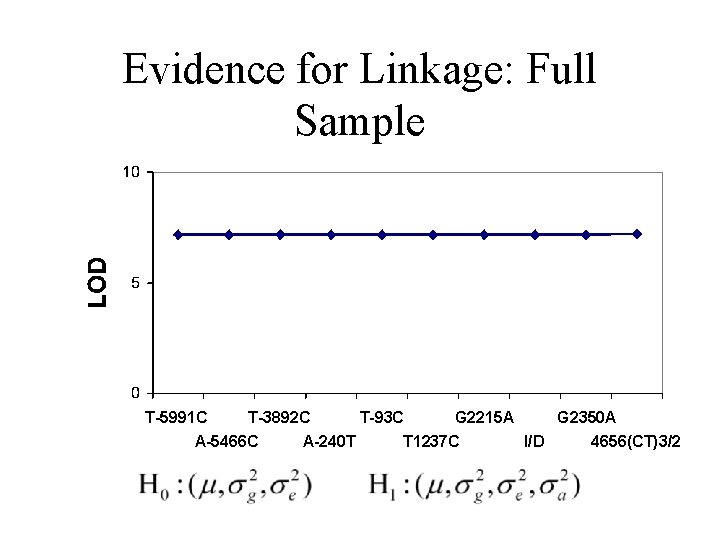
Evidence for Linkage: Full Sample T-5991 C T-3892 C A-5466 C A-240 T T-93 C G 2215 A T 1237 C G 2350 A I/D 4656(CT)3/2

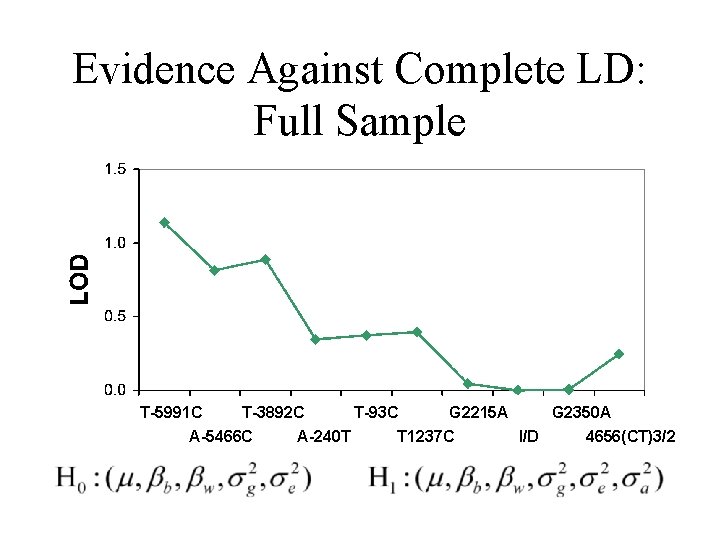
Evidence Against Complete LD: Full Sample T-5991 C T-3892 C A-5466 C A-240 T T-93 C G 2215 A T 1237 C G 2350 A I/D 4656(CT)3/2

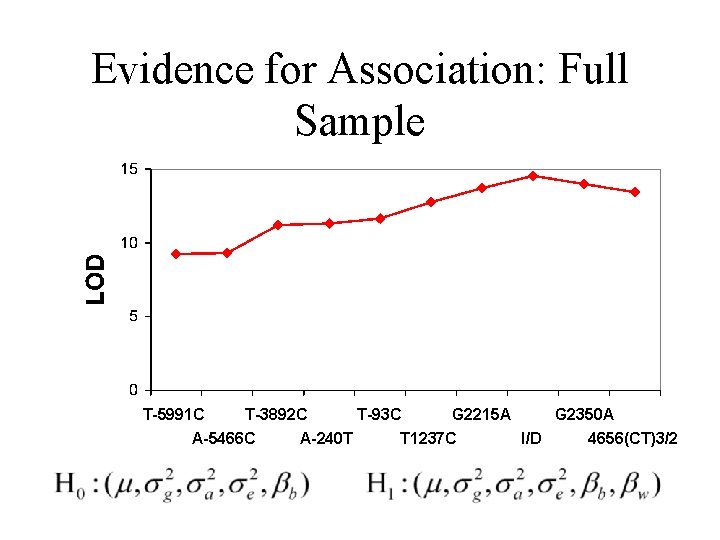
Evidence for Association: Full Sample T-5991 C T-3892 C A-5466 C A-240 T T-93 C G 2215 A T 1237 C G 2350 A I/D 4656(CT)3/2

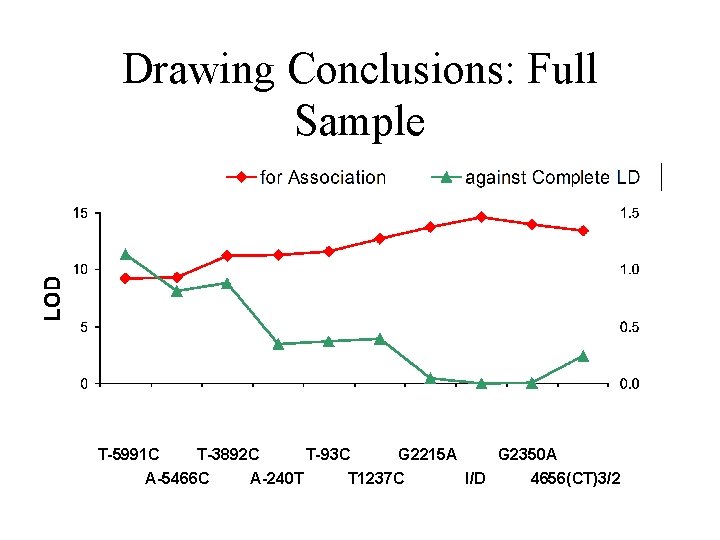
Drawing Conclusions: Full Sample T-5991 C T-3892 C T-93 C G 2215 A G 2350 A A-5466 C A-240 T T 1237 C I/D 4656(CT)3/2

ACE Example Summary • Agrees with haplotype analysis • Distinguishes complete and incomplete disequilibrium – Measure of distance for incomplete LD – Indicator of trait allele frequencies • Typical or fairy-tale?



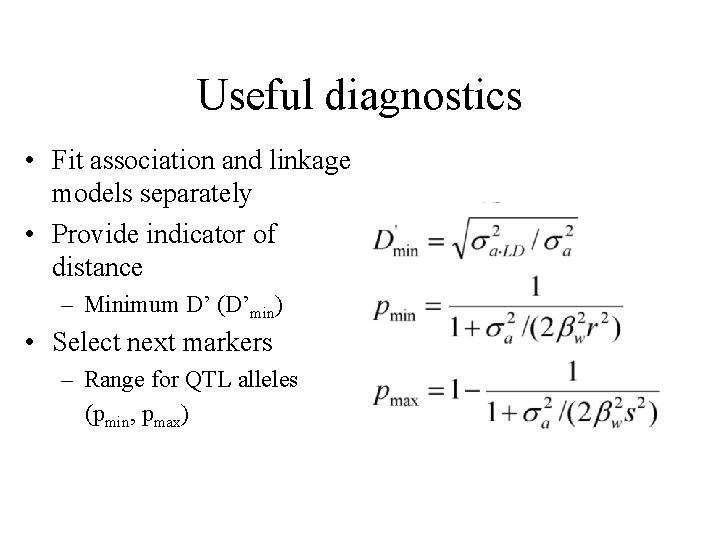
Useful diagnostics • Fit association and linkage models separately • Provide indicator of distance – Minimum D’ (D’min) • Select next markers – Range for QTL alleles (pmin, pmax)

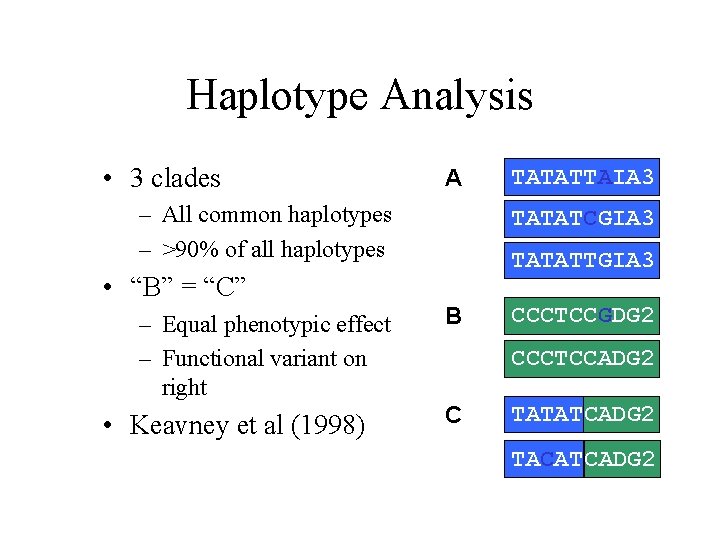
Haplotype Analysis • 3 clades A – All common haplotypes – >90% of all haplotypes TATATCGIA 3 TATATTGIA 3 • “B” = “C” – Equal phenotypic effect – Functional variant on right • Keavney et al (1998) TATATTAIA 3 B CCCTCCGDG 2 CCCTCCADG 2 C TATATCADG 2 TACATCADG 2


The Spielman TDT • Traditional case-control – Compare allele frequencies in two samples • Cases and controls must be one population • Heterozygous parents – Parental alleles are the study population – Population allele frequencies fixed • 50: 50, independent of original – Test for excess among affected offspring

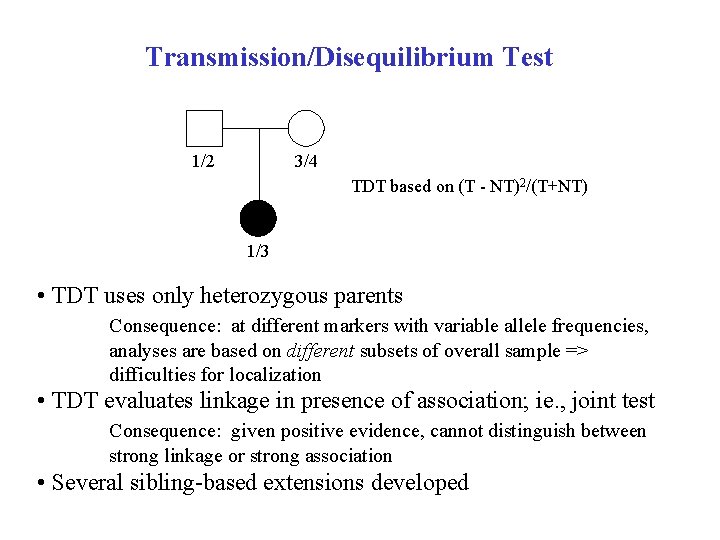
Transmission/Disequilibrium Test 1/2 3/4 TDT based on (T - NT)2/(T+NT) 1/3 • TDT uses only heterozygous parents Consequence: at different markers with variable allele frequencies, analyses are based on different subsets of overall sample => difficulties for localization • TDT evaluates linkage in presence of association; ie. , joint test Consequence: given positive evidence, cannot distinguish between strong linkage or strong association • Several sibling-based extensions developed

Family-based Association Methods for Quantitative Traits Primary aim: association test free of pop. sub-structure effects Allison, D. B. , AJHG, 1997 Rabinowitz, D. Hum Hered, 1997 Fulker, D. W. et al. AJHG, 1999 Elston, R. C. et al. AJHG, 1999 Allison, D. B. et al. AJHG, 1999 Abecasis, G. et al. AJHG 2000 Cardon, L. R. Hum Hered 2000 Monks, S. et al. abstract 1999 Selected parent-offspring trios Nuclear families Sib-pairs without parents General pedigrees (linkage) Sibships with/without parents (linkage) General pedigrees with/without parents Sib-pairs with Gx. E, epistatic interactions Nuclear families

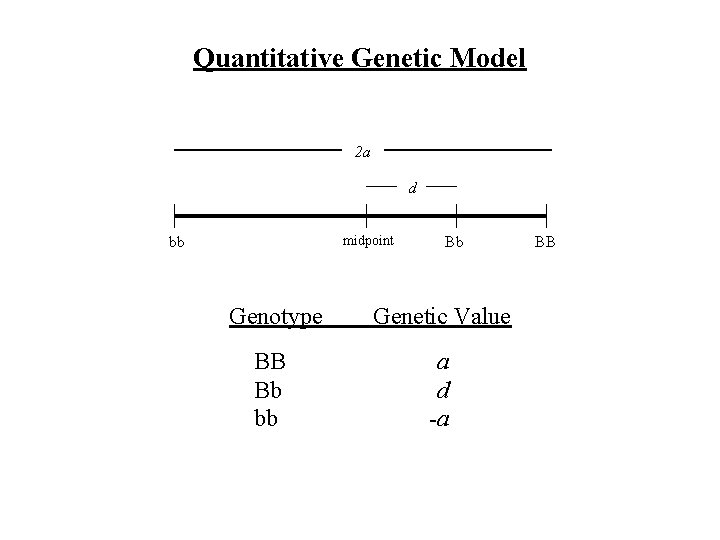
Quantitative Genetic Model 2 a d midpoint bb Bb Genotype Genetic Value BB Bb bb a d -a BB

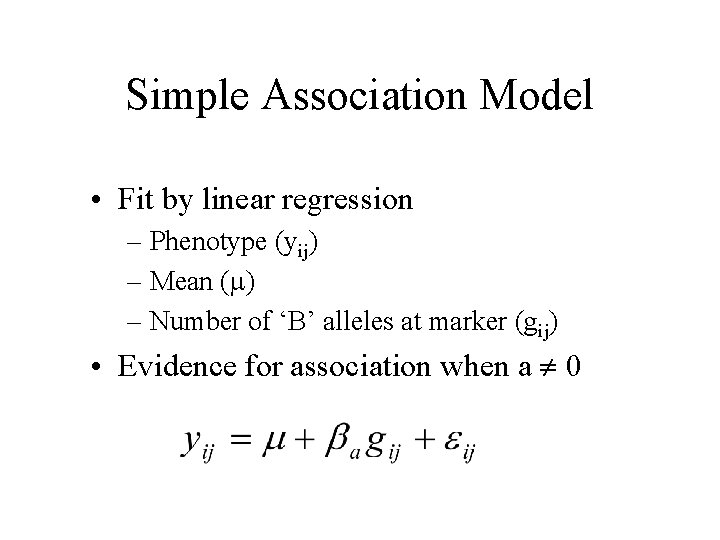
Simple Association Model • Fit by linear regression – Phenotype (yij) – Mean ( ) – Number of ‘B’ alleles at marker (gij) • Evidence for association when a 0

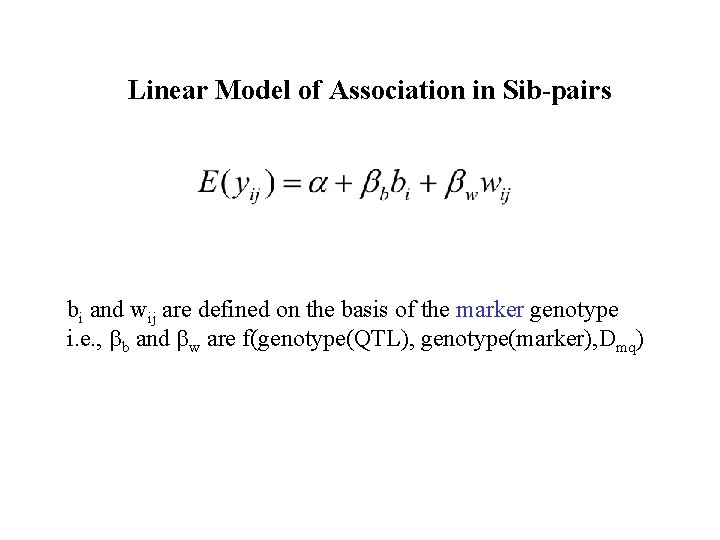
Linear Model of Association in Sib-pairs bi and wij are defined on the basis of the marker genotype i. e. , bb and bw are f(genotype(QTL), genotype(marker), Dmq)

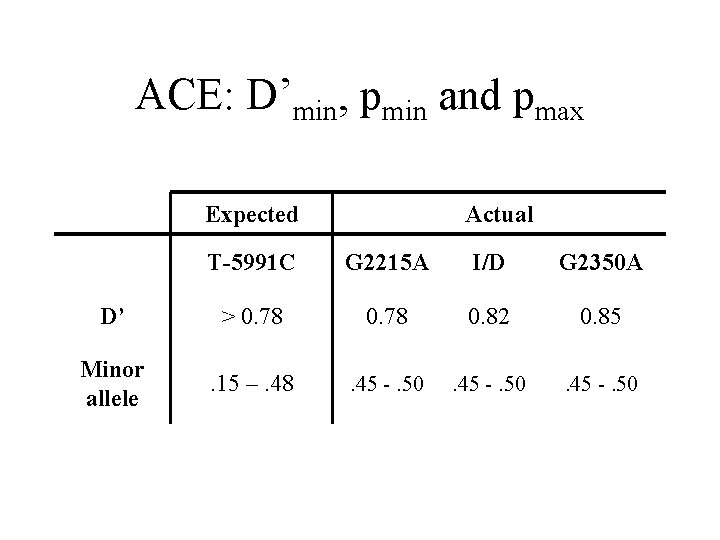
ACE: D’min, pmin and pmax Expected Actual T-5991 C G 2215 A I/D G 2350 A D’ > 0. 78 0. 82 0. 85 Minor allele . 15 –. 48 . 45 -. 50