SIMULATIONS OF PEPTIDE FOLDING and DYNAMICS Krzysztof Kuczera








![Folding: transition state w(n) = -k. Tln[P(n)] w(n) = -k. Tln[P(n)/C(n)] C(n) = 17!/n!(17 Folding: transition state w(n) = -k. Tln[P(n)] w(n) = -k. Tln[P(n)/C(n)] C(n) = 17!/n!(17](https://slidetodoc.com/presentation_image_h/9f5e33636deb2d48cbe9cd277e98be0e/image-17.jpg)




- Slides: 27

SIMULATIONS OF PEPTIDE FOLDING and DYNAMICS Krzysztof Kuczera Departments of Chemistry and Molecular Biosciences University of Kansas IMA Workshop Jan 14 -18, 2008

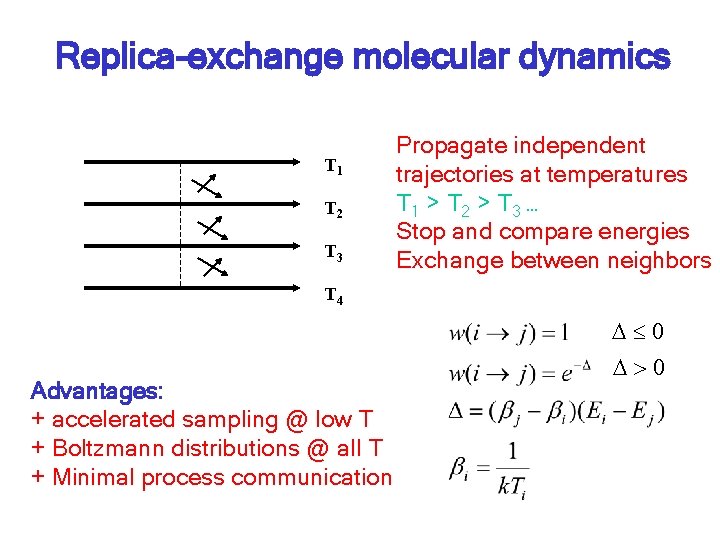
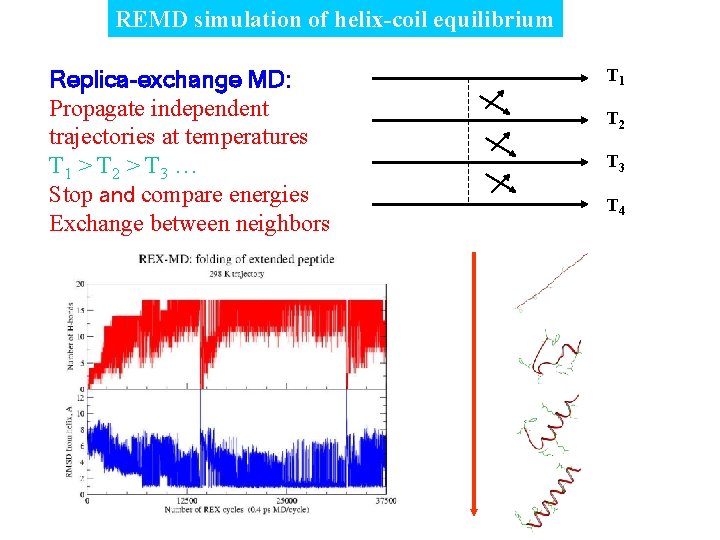
Replica-exchange molecular dynamics T 1 T 2 T 3 Propagate independent trajectories at temperatures T 1 > T 2 > T 3 … Stop and compare energies Exchange between neighbors T 4 Advantages: + accelerated sampling @ low T + Boltzmann distributions @ all T + Minimal process communication D 0 D>0

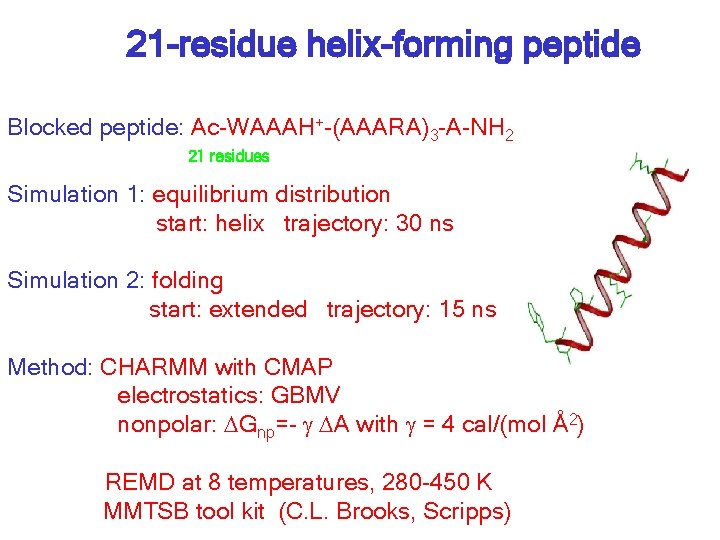
21 -residue helix-forming peptide Blocked peptide: Ac-WAAAH+-(AAARA)3 -A-NH 2 21 residues Simulation 1: equilibrium distribution start: helix trajectory: 30 ns Simulation 2: folding start: extended trajectory: 15 ns Method: CHARMM with CMAP electrostatics: GBMV nonpolar: DGnp=- g DA with g = 4 cal/(mol Å2) REMD at 8 temperatures, 280 -450 K MMTSB tool kit (C. L. Brooks, Scripps)





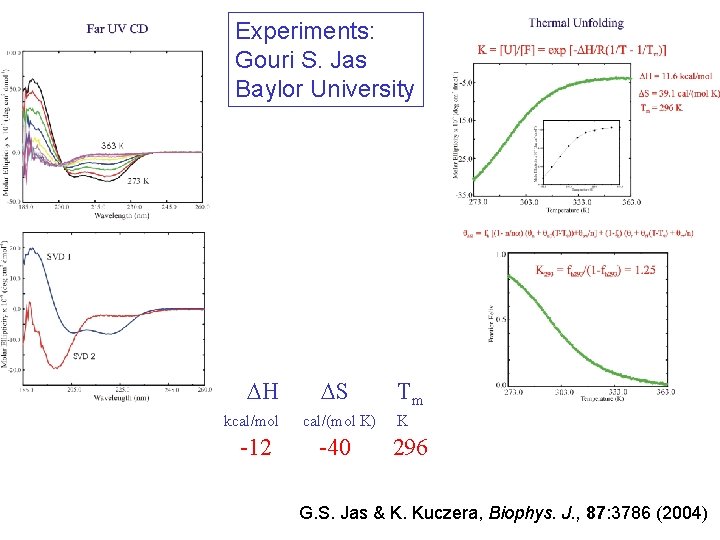
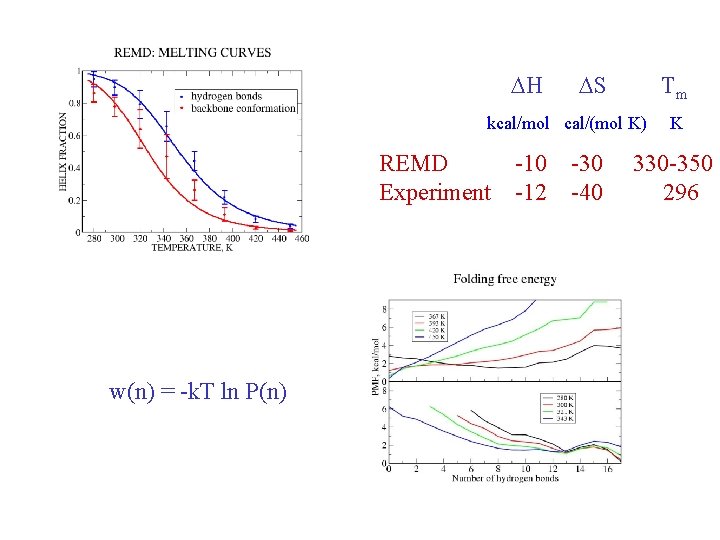
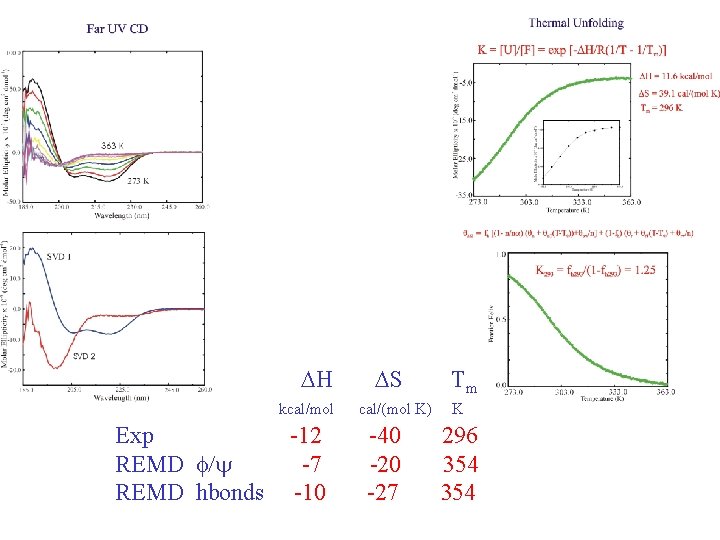
Experiments: Gouri S. Jas Baylor University DH kcal/mol -12 DS cal/(mol K) -40 Tm K 296 G. S. Jas & K. Kuczera, Biophys. J. , 87: 3786 (2004)

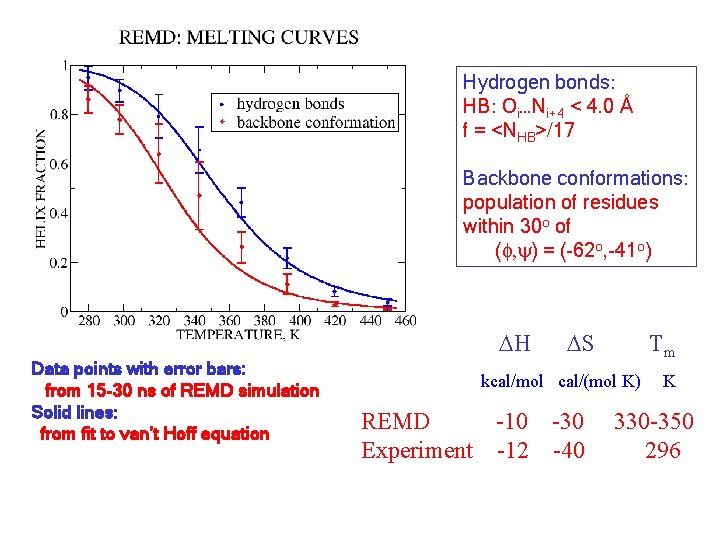
Hydrogen bonds: HB: Oi…Ni+4 < 4. 0 Å f = <NHB>/17 Backbone conformations: population of residues within 30 o of (f, y) = (-62 o, -41 o) DH Data points with error bars: from 15 -30 ns of REMD simulation Solid lines: from fit to van’t Hoff equation DS Tm kcal/mol cal/(mol K) REMD -10 -30 Experiment -12 -40 K 330 -350 296

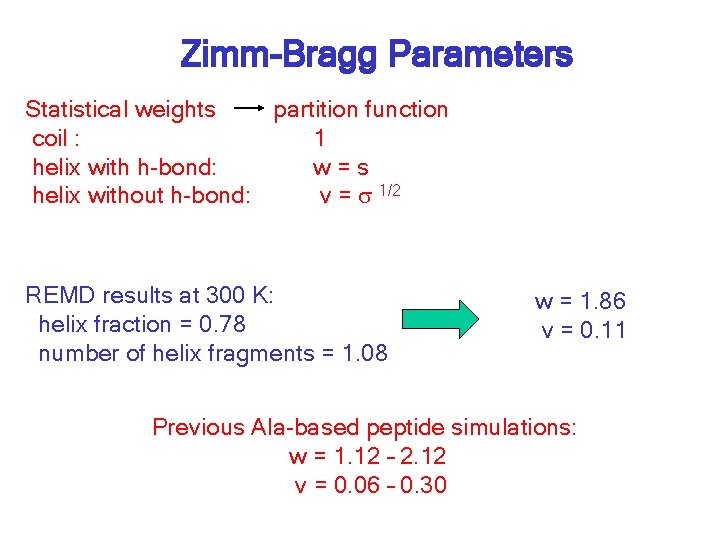
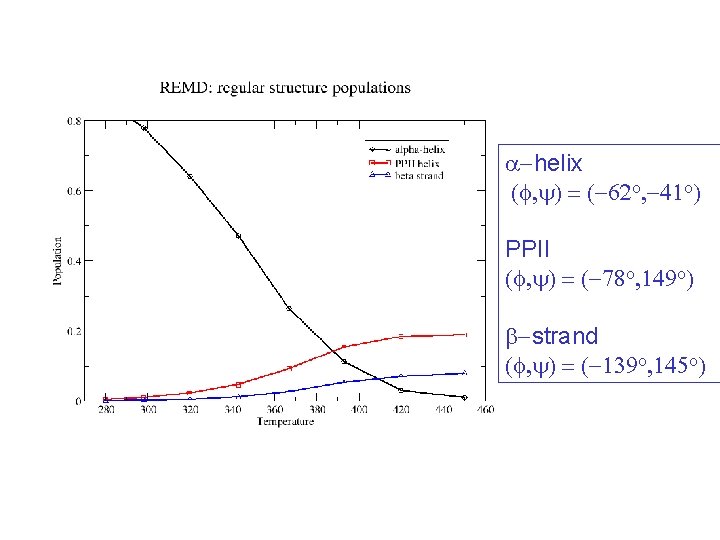
Zimm-Bragg Parameters Statistical weights partition function coil : 1 helix with h-bond: w=s helix without h-bond: v = s 1/2 REMD results at 300 K: helix fraction = 0. 78 number of helix fragments = 1. 08 w = 1. 86 v = 0. 11 Previous Ala-based peptide simulations: w = 1. 12 – 2. 12 v = 0. 06 – 0. 30



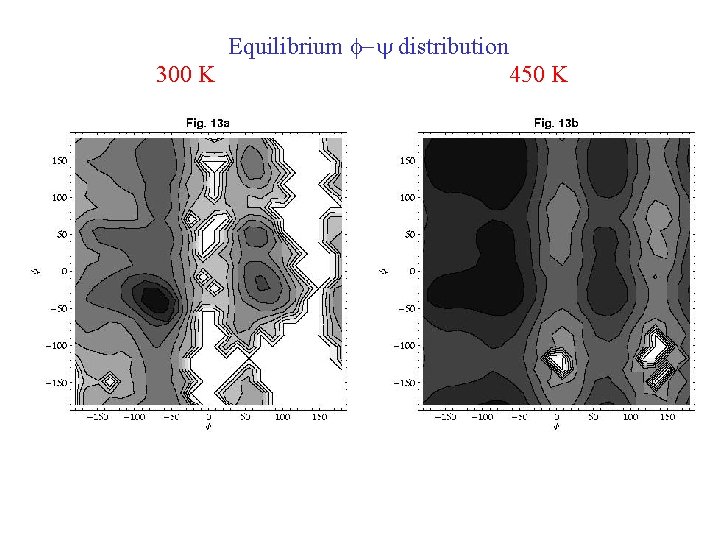
Equilibrium f-y distribution 300 K 450 K

a-helix (f, y) = (-62 o, -41 o) PPII (f, y) = (-78 o, 149 o) b-strand (f, y) = (-139 o, 145 o)


![Folding transition state wn k TlnPn wn k TlnPnCn Cn 17n17 Folding: transition state w(n) = -k. Tln[P(n)] w(n) = -k. Tln[P(n)/C(n)] C(n) = 17!/n!(17](https://slidetodoc.com/presentation_image_h/9f5e33636deb2d48cbe9cd277e98be0e/image-17.jpg)
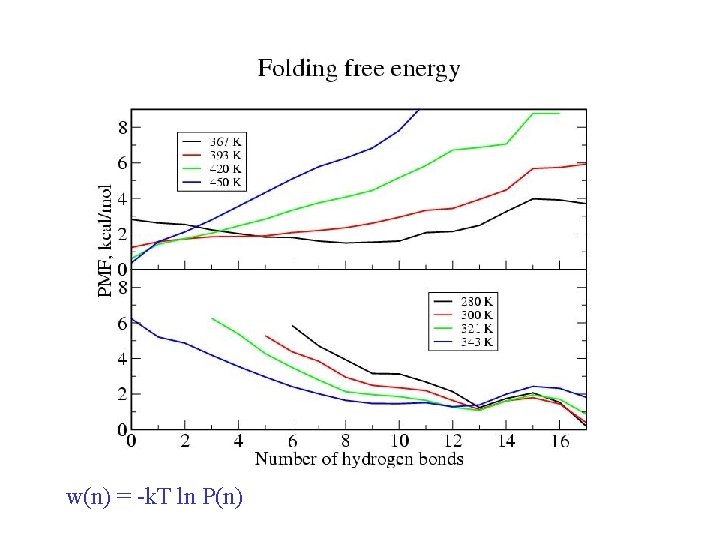
Folding: transition state w(n) = -k. Tln[P(n)] w(n) = -k. Tln[P(n)/C(n)] C(n) = 17!/n!(17 -n)!

Equilibrium REMD: conclusions • We have calculated an equilibrium melting curve for a helix-forming peptide with GB/SA model • Thermodynamics qualitatively correct Melting temperature exaggerated • Global free energy minimum = a-helix at low T = coil at high T • Microscopic information about helix unfolding - unfolding initiated at termini transition state ½ helix - formation of compact structures at low T - b and PPII structures at high T

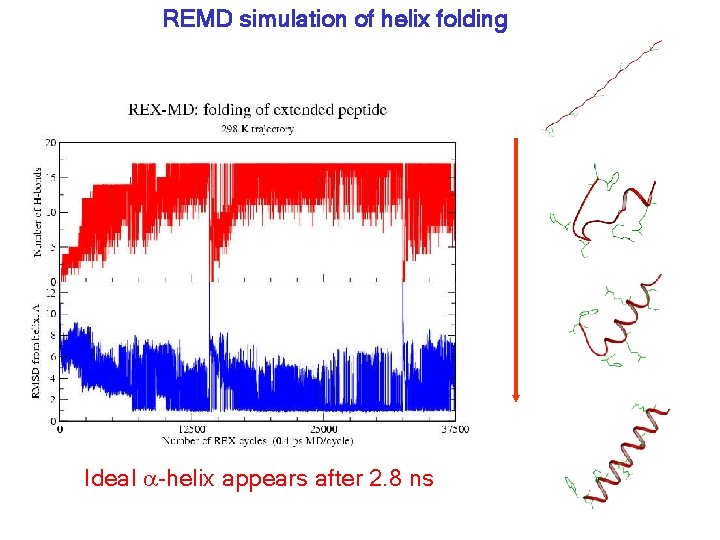
REMD simulation of helix folding Ideal a-helix appears after 2. 8 ns



Folding REMD: conclusions • a-helix structure found after ca. 3 ns simulation time • confirm that helix is global FE minimum, not a memory effect • folding in room T replica is sequential • structures and properties sampled are similar to trajectory starting from helix • conformational sampling acceleration of REX-MD over direct MD is ca. 100 overall (ca. 12 per CPU) [3 ns folding in REMD vs. 300 ns exp time scale]

Acknowledgments • Gerald Lushington, University of Kansas Molecular Modeling and Graphics Laboratory – Athlon cluster • Michael Feig, John Caranicolas and Charles L. Brooks III MMTSB Tool Set (2001), The Scripps Research Institute • Gouri S. Jas, Baylor University - experiments • ACS PRF $$$

DH DS Tm kcal/mol cal/(mol K) REMD -10 -30 Experiment -12 -40 w(n) = -k. T ln P(n) K 330 -350 296

REMD simulation of helix-coil equilibrium Replica-exchange MD: Propagate independent trajectories at temperatures T 1 > T 2 > T 3 … Stop and compare energies Exchange between neighbors T 1 T 2 T 3 T 4

DH kcal/mol Exp REMD f/y REMD hbonds -12 -7 -10 DS cal/(mol K) -40 -27 Tm K 296 354

w(n) = -k. T ln P(n)
 Krzysztof kuczera
Krzysztof kuczera Yes or no
Yes or no Clinical simulations in nursing education
Clinical simulations in nursing education Chris harding simulations
Chris harding simulations Tcad simulations
Tcad simulations World history simulations
World history simulations Simulations for solid state physics
Simulations for solid state physics Www.irs.gov/app/understanding taxes/student/simulations.jsp
Www.irs.gov/app/understanding taxes/student/simulations.jsp Pinpoint simulations 777
Pinpoint simulations 777 Baton simulations
Baton simulations Rigid planar
Rigid planar Peptide bonds in primary structure of protein
Peptide bonds in primary structure of protein How many amino acids are there
How many amino acids are there Salt bond cosmetology
Salt bond cosmetology Simple proteins
Simple proteins Elevated c peptide
Elevated c peptide Pulmonary hypertension differential diagnosis
Pulmonary hypertension differential diagnosis Low c peptide
Low c peptide Bremelanotide australia
Bremelanotide australia Peptide bond importance
Peptide bond importance Number of peptide bonds
Number of peptide bonds Renal plasma flow
Renal plasma flow Mascot peptide mass fingerprint
Mascot peptide mass fingerprint Atrial natriuretic peptide
Atrial natriuretic peptide What biological molecules contain peptide bonds
What biological molecules contain peptide bonds Peptide identification
Peptide identification Peptide identification
Peptide identification Primary secondary tertiary quaternary
Primary secondary tertiary quaternary