Aggressive Enumeration of Peptide Sequences for MSMS Peptide



![EST Peptides • Frame 1 translation CTTKFCDYGKA PGAEEYAQQDV LKKSYSKAFTL TISALFVTPKT TGA[QPRL]VELSEQQ LQL[S*LW]PSDVDKL SPTD[IKMNSRT] 23 EST Peptides • Frame 1 translation CTTKFCDYGKA PGAEEYAQQDV LKKSYSKAFTL TISALFVTPKT TGA[QPRL]VELSEQQ LQL[S*LW]PSDVDKL SPTD[IKMNSRT] 23](https://slidetodoc.com/presentation_image_h2/c354de8b84449b7a0655ef8da70d17d1/image-23.jpg)

- Slides: 45

Aggressive Enumeration of Peptide Sequences for MS/MS Peptide Identification Nathan Edwards Center for Bioinformatics and Computational Biology

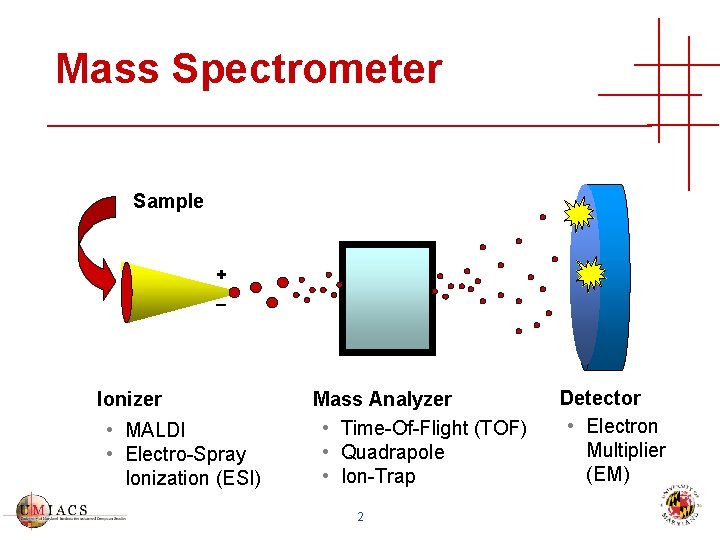
Mass Spectrometer Sample + _ Ionizer • MALDI • Electro-Spray Ionization (ESI) Mass Analyzer • Time-Of-Flight (TOF) • Quadrapole • Ion-Trap 2 Detector • Electron Multiplier (EM)

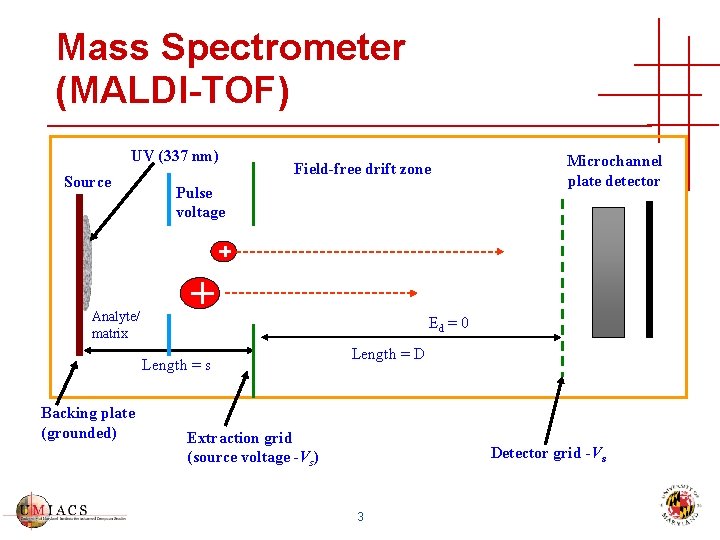
Mass Spectrometer (MALDI-TOF) UV (337 nm) Source Field-free drift zone Pulse voltage Analyte/ matrix Ed = 0 Length = s Backing plate (grounded) Microchannel plate detector Length = D Extraction grid (source voltage -Vs) Detector grid -Vs 3

Mass is fundamental 4

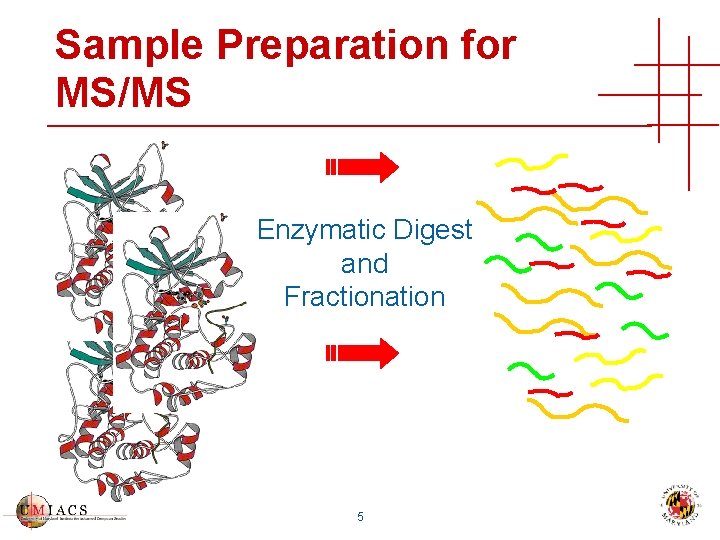
Sample Preparation for MS/MS Enzymatic Digest and Fractionation 5

Single Stage MS MS 6

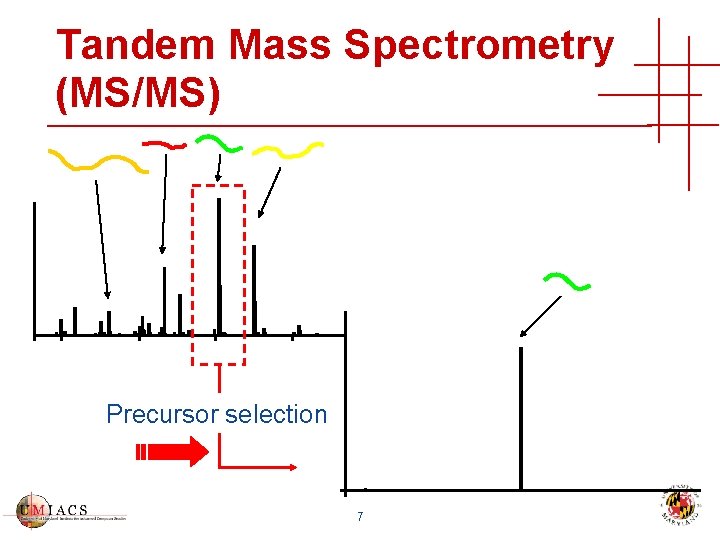
Tandem Mass Spectrometry (MS/MS) Precursor selection 7

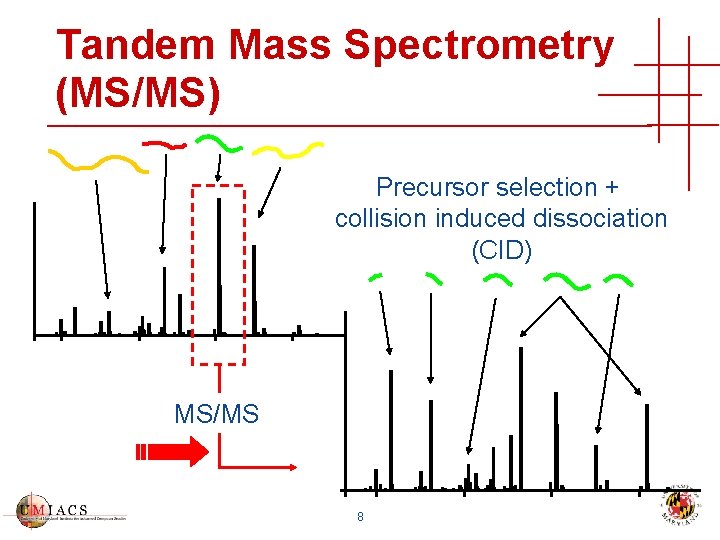
Tandem Mass Spectrometry (MS/MS) Precursor selection + collision induced dissociation (CID) MS/MS 8

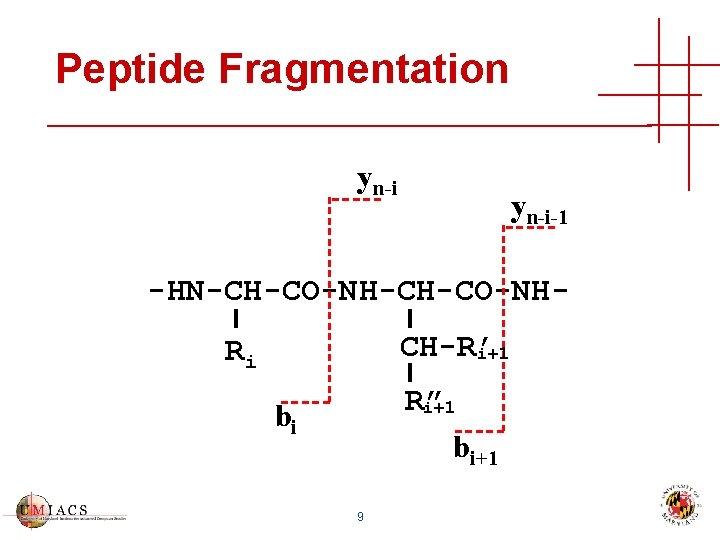
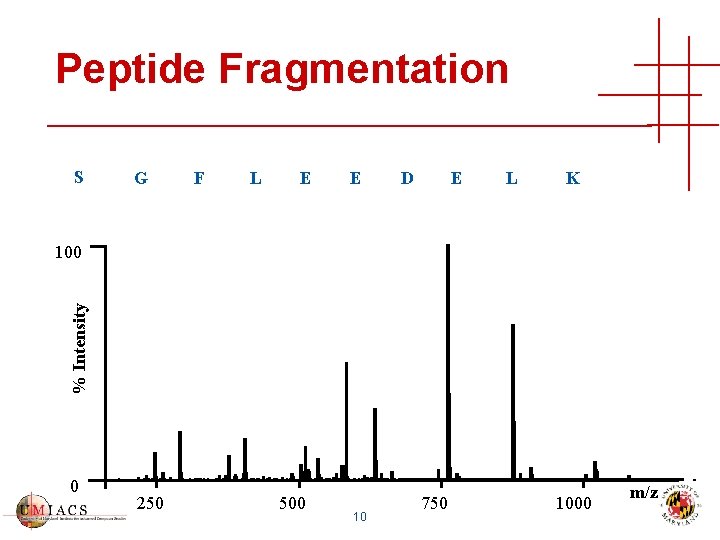
Peptide Fragmentation yn-i-1 -HN-CH-CO-NHCH-R’ i+1 Ri R” i+1 bi 9

Peptide Fragmentation S G F L E E D E L K % Intensity 100 0 250 500 10 750 1000 m/z

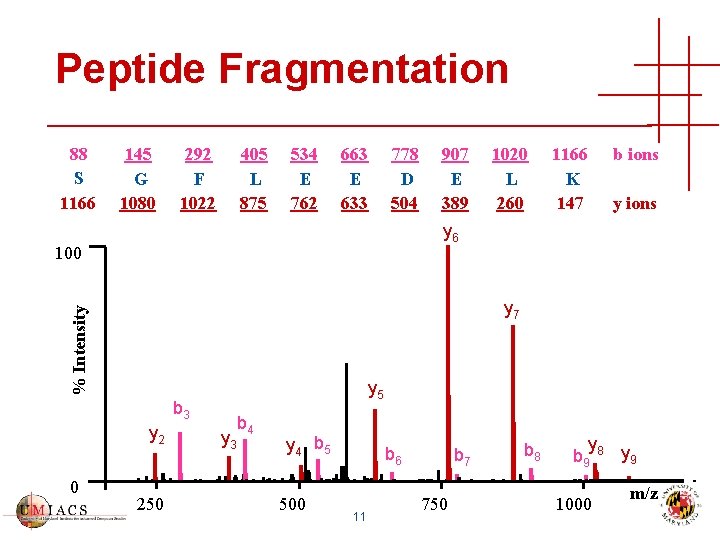
Peptide Fragmentation 88 S 1166 145 G 1080 292 F 1022 405 L 875 534 E 762 663 E 633 778 D 504 1020 L 260 1166 K 147 b ions y 6 100 % Intensity y 7 y 5 b 3 y 2 0 907 E 389 250 y 3 b 4 y 4 b 5 500 b 6 11 b 7 750 b 8 y b 9 8 y 9 1000 m/z

MS/MS Search Engines • Fail when peptides are missing from sequence database • Protein sequence databases serve many masters • Full length protein sequences not needed for MS/MS • Explicit variant enumeration is needed for MS/MS • Much peptide sequence information is lost, inaccessible, or not integrated • Protein isoforms, sequence variants, SNPs, alternate splice forms, ESTs • Some peptides are more interesting than others • Protein identification is only part of the story 12

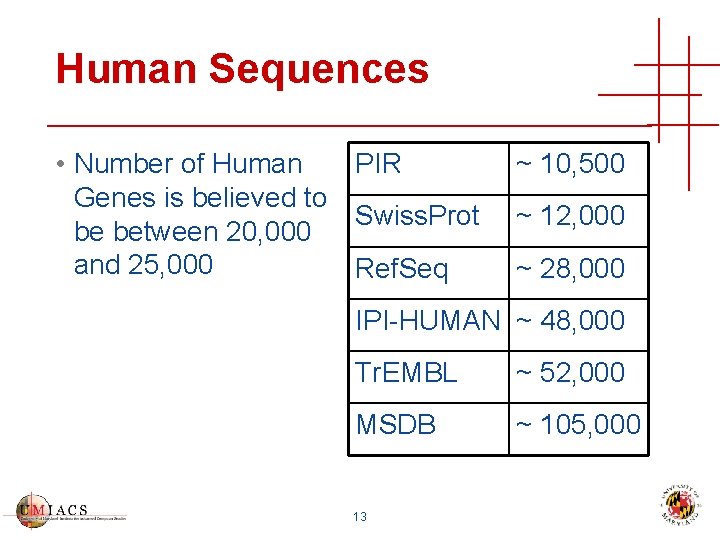
Human Sequences PIR • Number of Human Genes is believed to Swiss. Prot be between 20, 000 and 25, 000 Ref. Seq ~ 10, 500 ~ 12, 000 ~ 28, 000 IPI-HUMAN ~ 48, 000 Tr. EMBL ~ 52, 000 MSDB ~ 105, 000 13

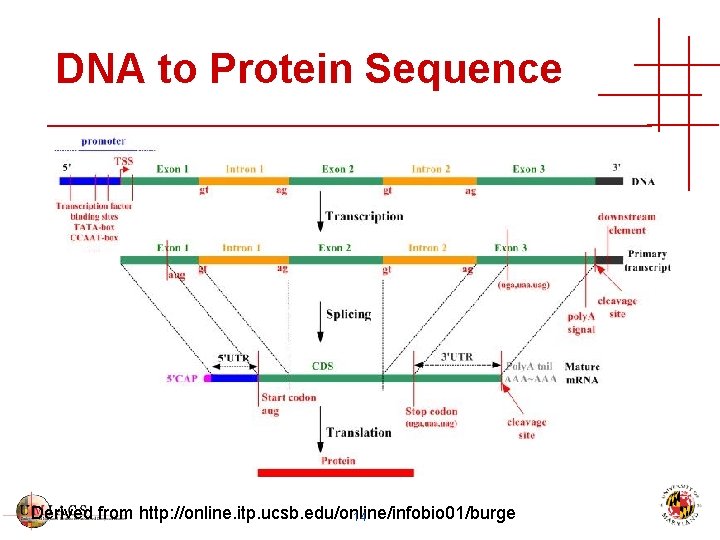
DNA to Protein Sequence Derived from http: //online. itp. ucsb. edu/online/infobio 01/burge 14

UCSC Genome Brower 15

Genomic Peptide Sequences • Many putative peptide sequences never become “protein” sequences • Genomic DNA, • Refseq m. RNA, ESTs • SNP/Polymorphism databases • Variant records in Swiss. Prot • Genomic annotation seeks “full length” genes and proteins 16

Genomic Peptide Sequences • Genomic DNA • Exons & introns, 6 frames, large (3 Gb → 6 Gb) • Refseq m. RNA • No introns, 3 frames, small (36 Mb → 36 Mb) • Most protein sequences already represented in sequence databases • ESTs • No introns, 6 frames, large (3 Gb → 6 Gb) • Used by gene & alternative splicing pipelines • Highly redundant, nucleotide error rate ~ 1% 17

“Novel” Peptide 18

Novel peptide 19

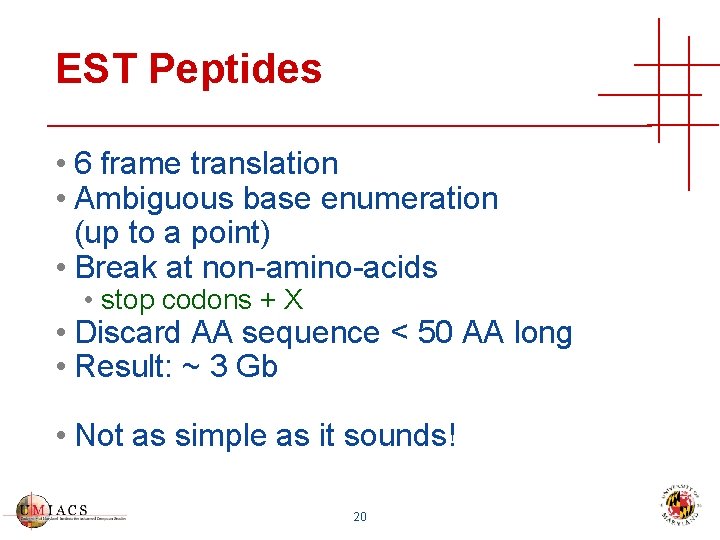
EST Peptides • 6 frame translation • Ambiguous base enumeration (up to a point) • Break at non-amino-acids • stop codons + X • Discard AA sequence < 50 AA long • Result: ~ 3 Gb • Not as simple as it sounds! 20

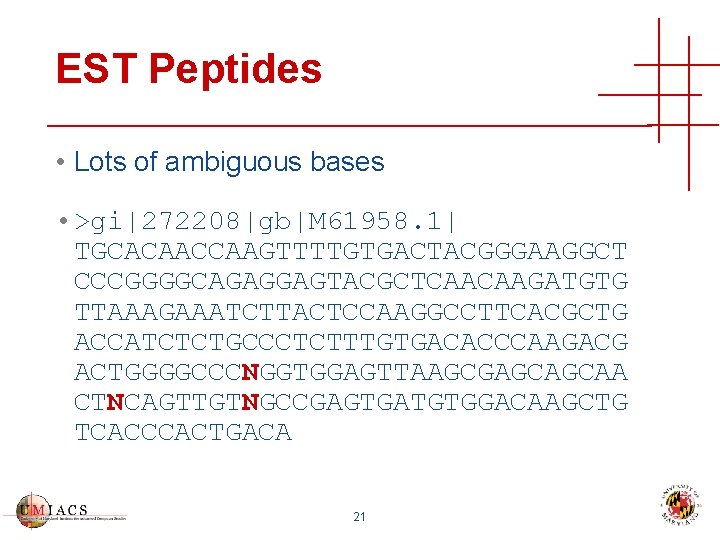
EST Peptides • Lots of ambiguous bases • >gi|272208|gb|M 61958. 1| TGCACAACCAAGTTTTGTGACTACGGGAAGGCT CCCGGGGCAGAGGAGTACGCTCAACAAGATGTG TTAAAGAAATCTTACTCCAAGGCCTTCACGCTG ACCATCTCTGCCCTCTTTGTGACACCCAAGACG ACTGGGGCCCNGGTGGAGTTAAGCGAGCAGCAA CTNCAGTTGTNGCCGAGTGATGTGGACAAGCTG TCACCCACTGACA 21

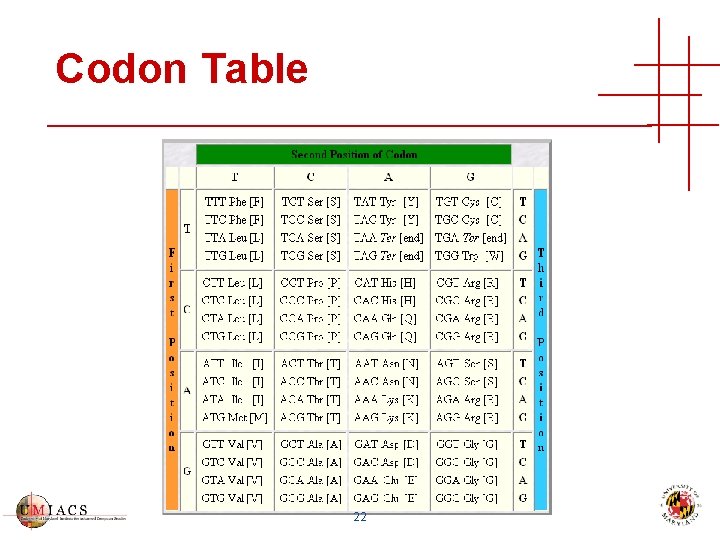
Codon Table 22
![EST Peptides Frame 1 translation CTTKFCDYGKA PGAEEYAQQDV LKKSYSKAFTL TISALFVTPKT TGAQPRLVELSEQQ LQLSLWPSDVDKL SPTDIKMNSRT 23 EST Peptides • Frame 1 translation CTTKFCDYGKA PGAEEYAQQDV LKKSYSKAFTL TISALFVTPKT TGA[QPRL]VELSEQQ LQL[S*LW]PSDVDKL SPTD[IKMNSRT] 23](https://slidetodoc.com/presentation_image_h2/c354de8b84449b7a0655ef8da70d17d1/image-23.jpg)
EST Peptides • Frame 1 translation CTTKFCDYGKA PGAEEYAQQDV LKKSYSKAFTL TISALFVTPKT TGA[QPRL]VELSEQQ LQL[S*LW]PSDVDKL SPTD[IKMNSRT] 23

Correcting EST Sequence • Align ESTs to genome • Use aligned genomic sequence • Must get splice sites right! • 6 frame translation • Break at non-amino-acids • stop codons + X • Discard AA sequence < 50 AA long • Result: ~ 1 Gb 24

Genomic Coding Sequence • Use Genscan to predict exons • Use very low probability threshold • Alternative exons option • No need for translation (35 Mb) 25

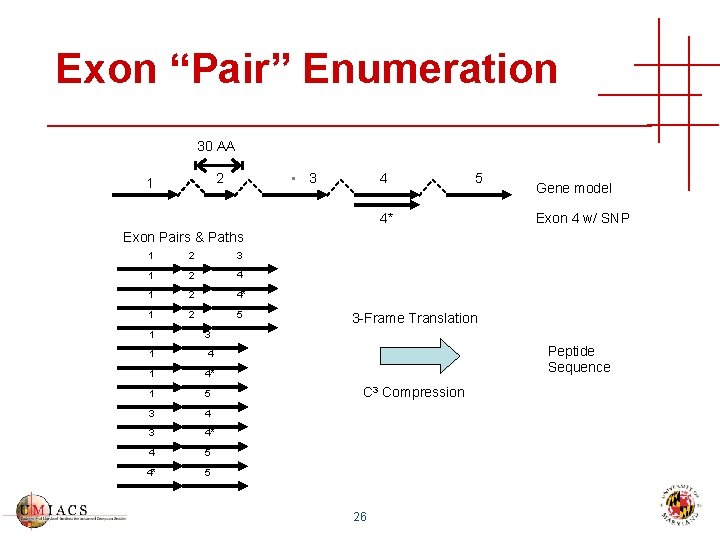
Exon “Pair” Enumeration 30 AA • 3 2 1 4 5 4* Gene model Exon 4 w/ SNP Exon Pairs & Paths 1 2 3 1 2 4* 1 2 5 1 3 1 4* 1 5 3 4* 4 5 4* 5 3 -Frame Translation Peptide Sequence C 3 Compression 26

Peptide Candidates • Parent ion • Typically < 3000 Da • Tryptic Peptides • Cut at K or R • Search engines • Don’t handle > 4+ well • Long peptides don’t fragment well • # of distinct 30 -mers upper bounds total peptide content 27

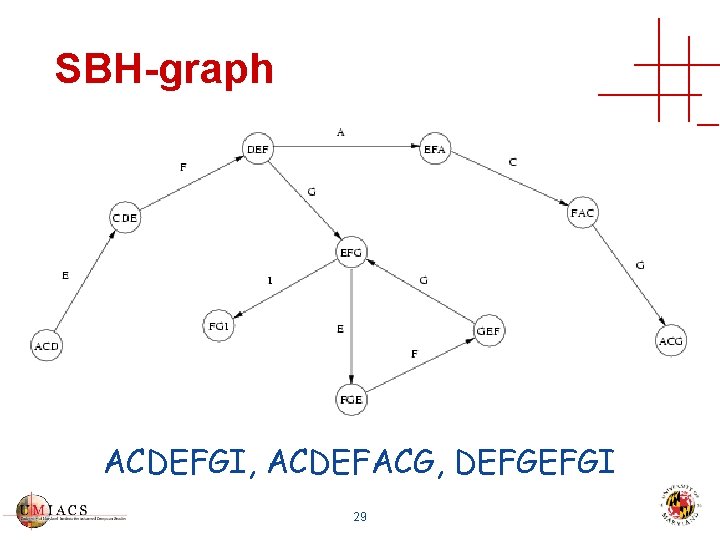
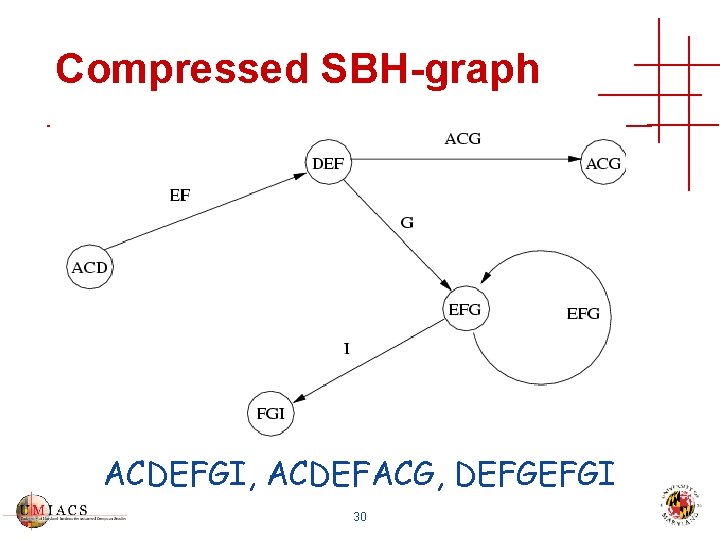
Sequence Database Compression Construct sequence database that is • Complete • All 30 -mers are present • Correct • No other 30 -mers are present • Compact • No 30 -mer is present more than once 28

SBH-graph ACDEFGI, ACDEFACG, DEFGEFGI 29

Compressed SBH-graph ACDEFGI, ACDEFACG, DEFGEFGI 30

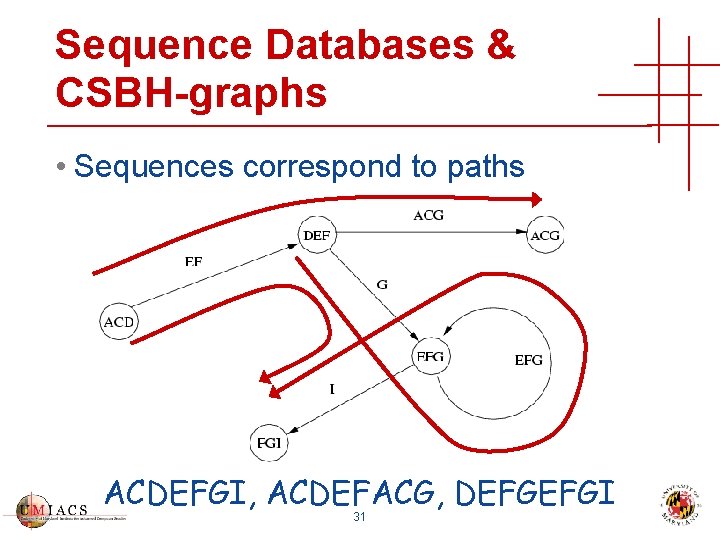
Sequence Databases & CSBH-graphs • Sequences correspond to paths ACDEFGI, ACDEFACG, DEFGEFGI 31

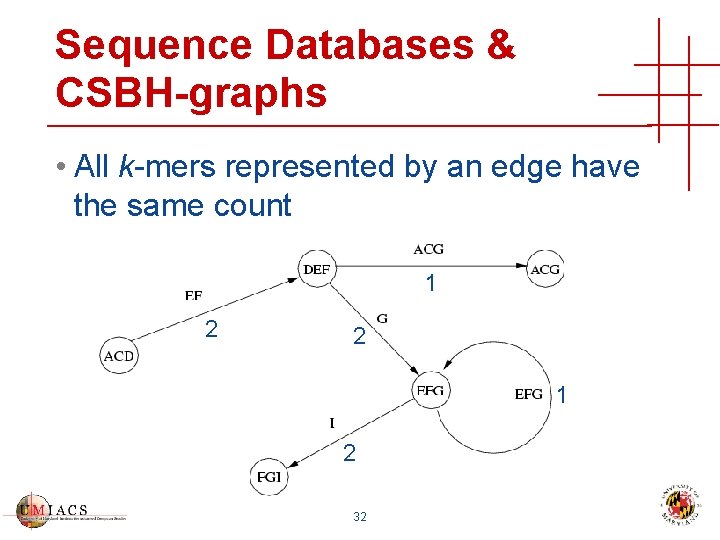
Sequence Databases & CSBH-graphs • All k-mers represented by an edge have the same count 1 2 2 1 2 32

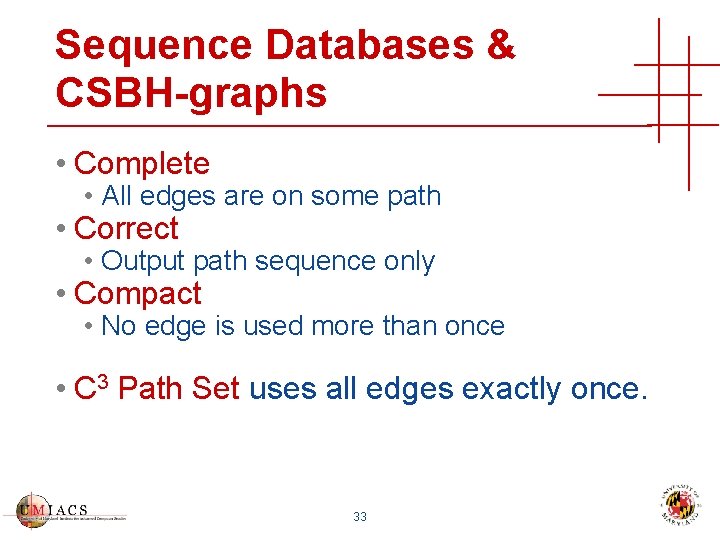
Sequence Databases & CSBH-graphs • Complete • All edges are on some path • Correct • Output path sequence only • Compact • No edge is used more than once • C 3 Path Set uses all edges exactly once. 33

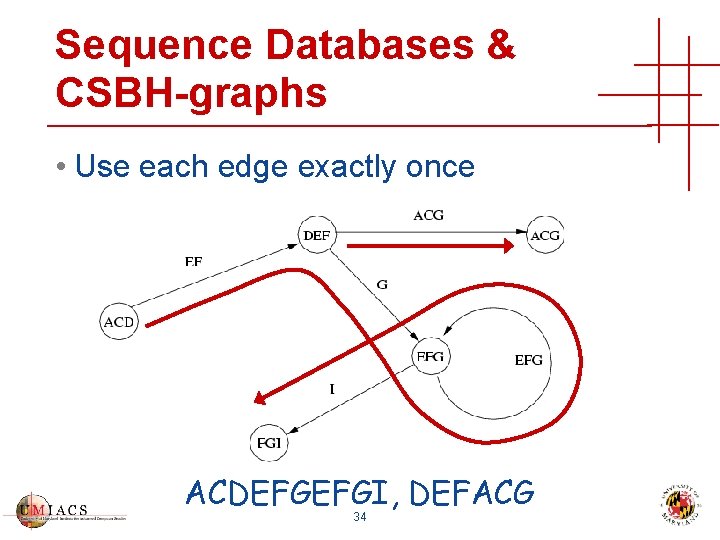
Sequence Databases & CSBH-graphs • Use each edge exactly once ACDEFGEFGI, DEFACG 34

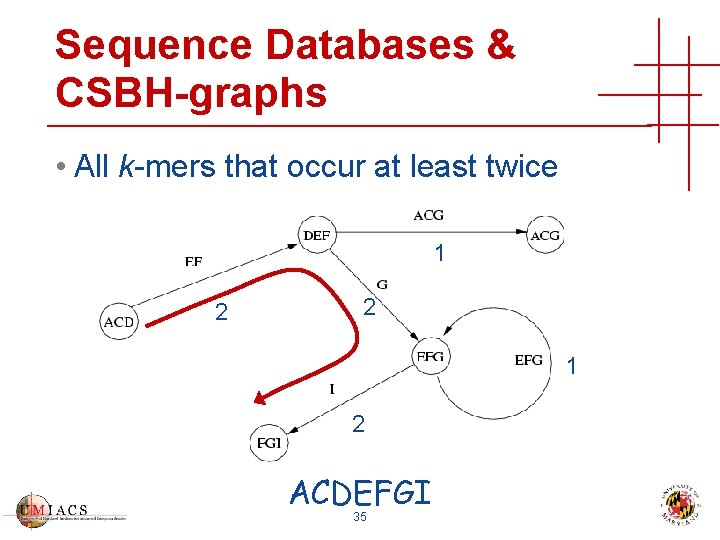
Sequence Databases & CSBH-graphs • All k-mers that occur at least twice 1 2 2 1 2 ACDEFGI 35

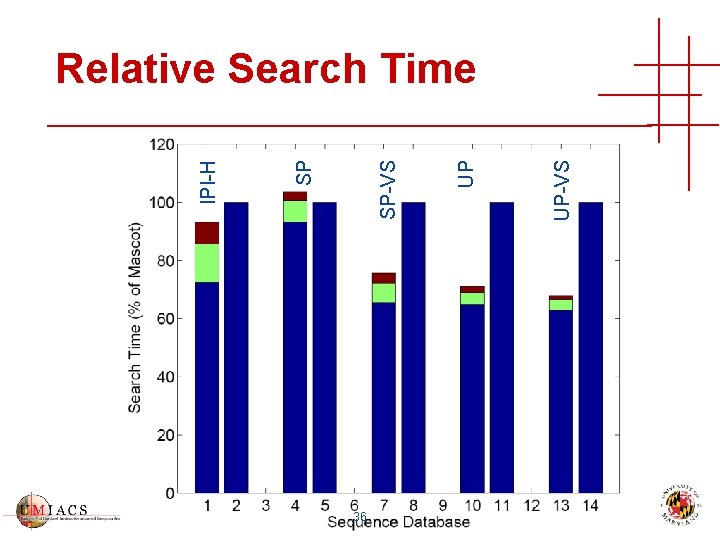
36 UP-VS UP SP-VS SP IPI-H Relative Search Time

More Sensitive Peptide ID • Significances, p-values, Expect values • Normalize for number of trials • Blast: • Size of sequence database • Mascot etc. : • Number of peptides scored against each spectrum • Redundant peptide sequences increase the number of trials, artificially. • Trials are not independent! • Less redundancy results in a better significance estimate 37

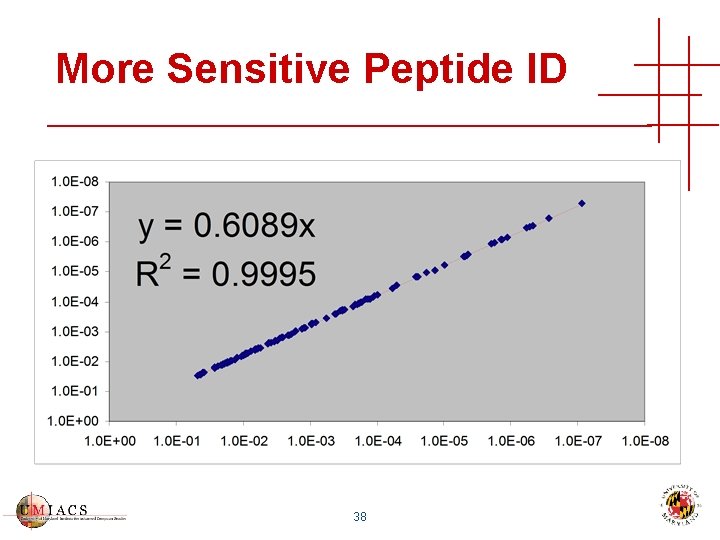
More Sensitive Peptide ID 38

Human Peptide Sequences • EST enumeration • 30 -mers must occur at least twice • EST corrections • Genscan exons • Uncompressed size: ~ 4. 5 Gb • Compressed size: ~ 263 Mb 39

Infrastructure • X!Tandem open source search engine • Configured to search aggressive peptide enumeration (human) • Web interface for browsing results • Integrated with condor • Results stored in My. SQL database • Over 3 million publicly available MS/MS spectra from human samples 40

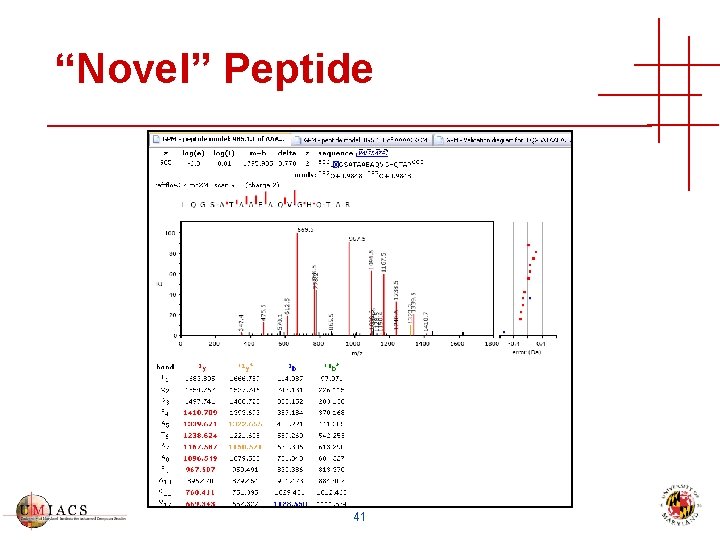
“Novel” Peptide 41

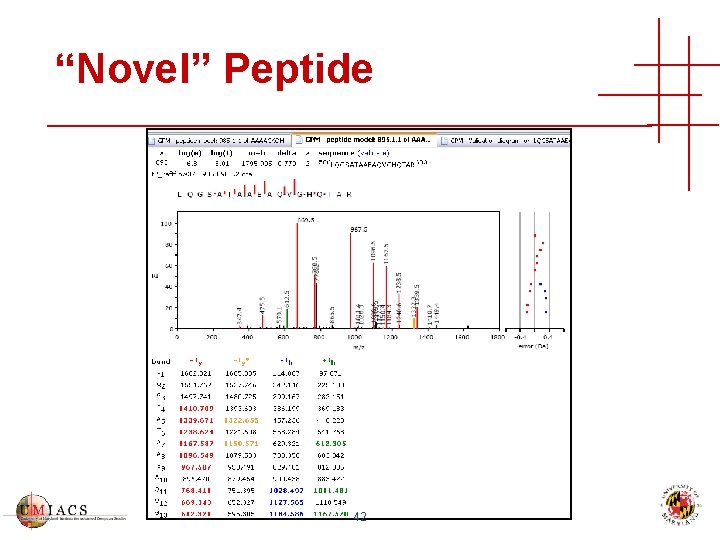
“Novel” Peptide 42

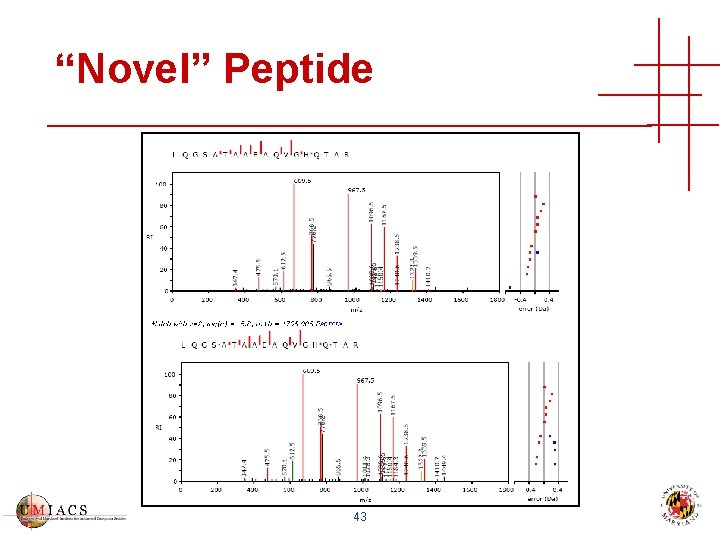
“Novel” Peptide 43

Ongoing work • Integrate SNPs and exon pairs • Get (lots) more spectra! • Solve the reverse mapping problem • Where did this peptide come from? • What protein does this peptide represent? 44

Thanks • Informatics Research @ ABI & Celera • Ross Lippert, Clark Mobarry, Bjarni Halldorsson • UMIACS @ University of Maryland, CP • V. S. Subrahmanian, Fritz Mc. Call, Doan Pham • Fenselau Lab @ UM, CP • CS @ University of Maryland, CP • Chau-Wen Tseng, Xue Wu 45