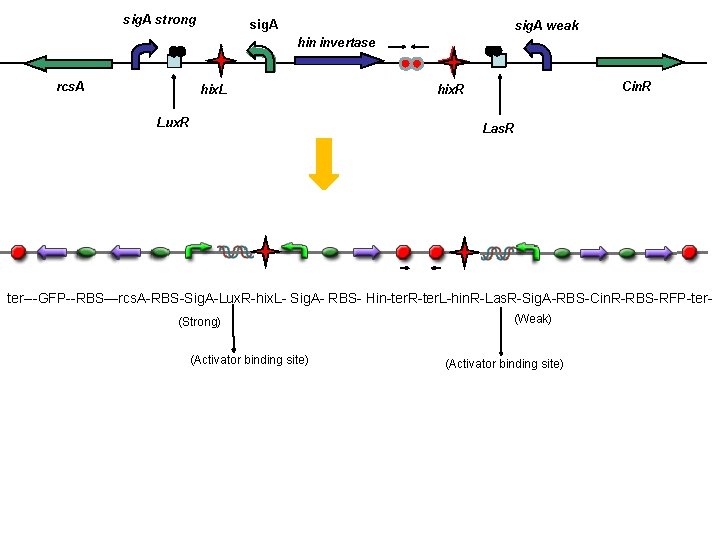
sig A strong sig A weak hin invertase








- Slides: 15


sig. A strong sig. A weak hin invertase rcs. A hix. L Lux. R Cin. R hix. R Las. R ter---GFP--RBS—rcs. A-RBS-Sig. A-Lux. R-hix. L- Sig. A- RBS- Hin-ter. R-ter. L-hin. R-Las. R-Sig. A-RBS-Cin. R-RBS-RFP-ter(Strong) (Activator binding site) (Weak) (Activator binding site)

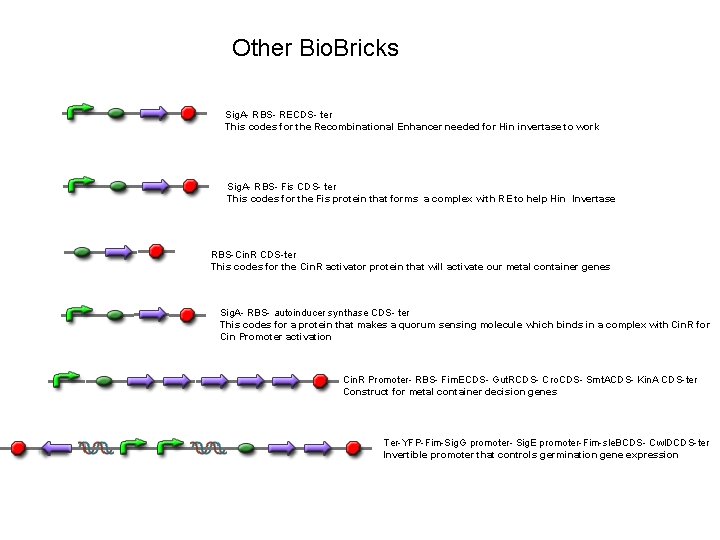
Other Bio. Bricks Sig. A- RBS- RECDS- ter This codes for the Recombinational Enhancer needed for Hin invertase to work Sig. A- RBS- Fis CDS- ter This codes for the Fis protein that forms a complex with RE to help Hin Invertase RBS-Cin. R CDS-ter This codes for the Cin. R activator protein that will activate our metal container genes Sig. A- RBS- autoinducer synthase CDS- ter This codes for a protein that makes a quorum sensing molecule which binds in a complex with Cin. R for Cin Promoter activation Cin. R Promoter- RBS- Fim. ECDS- Gut. RCDS- Cro. CDS- Smt. ACDS- Kin. A CDS-ter Construct for metal container decision genes Ter-YFP-Fim-Sig. G promoter- Sig. E promoter-Fim-sle. BCDS- Cwl. DCDS-ter Invertible promoter that controls germination gene expression

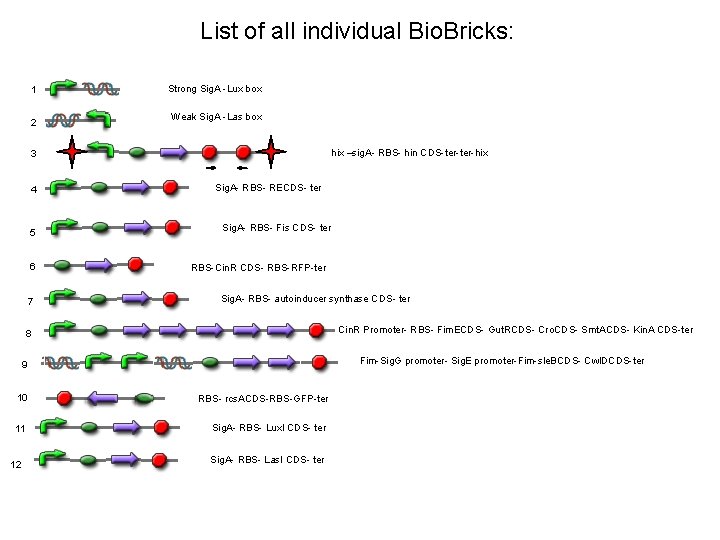
List of all individual Bio. Bricks: 1 2 Strong Sig. A -Lux box Weak Sig. A -Las box hix –sig. A- RBS- hin CDS-ter-hix 3 4 5 6 7 Sig. A- RBS- RECDS- ter Sig. A- RBS- Fis CDS- ter RBS-Cin. R CDS- RBS-RFP-ter Sig. A- RBS- autoinducer synthase CDS- ter Cin. R Promoter- RBS- Fim. ECDS- Gut. RCDS- Cro. CDS- Smt. ACDS- Kin. A CDS-ter 8 Fim-Sig. G promoter- Sig. E promoter-Fim-sle. BCDS- Cwl. DCDS-ter 9 10 RBS- rcs. ACDS-RBS-GFP-ter 11 Sig. A- RBS- Lux. I CDS- ter 12 Sig. A- RBS- Las. I CDS- ter

Part sequences 1. Sig. A from promoter library- Lux box consensus from Antunes et al. 2008 2. Sig. A from promoter library- Las. R-binding seq 3. Left hix (Bba_S 03383)- sig. A- RBS (BBa_K 090505)- Hin+LVA (BBa_J 31001)- ter-Right Hix (BBa_S 03384) 4. Sig. A from promoter library- RBS (BBa_K 090505)- Recombinational enhancer (BBa_J 3101)-ter 5. Sig. A from promoter library- RBS (BBa_K 090505)- Fis Protein (http: //www. ncbi. nlm. nih. gov/nuccore/242375837? from=3271637&to=3271933&report=gbwithparts)-ter RBS (BBa_K 090505)- Cin. R CDS (BBa_C 0077)-ter 6. 7. Sig. A- RBS (BBa_K 090505)- Autoinducer synthase CDS (BBa_C 0076)-ter 8. Cin. R promoter (BBa_R 0077)- RBS (BBa_K 090505)- Cro CDS (Roberts 1977 paper)-Smta CDS (brick. doc)kin. A CDS (http: //www. ncbi. nlm. nih. gov/nuccore/2632216? from=4382&to=6202&report=gbwithparts)- Fim. E CDS (http: //www. uniprot. org/uniprot/P 0 ADH 7. fasta)-ter 9. Fimsite (Mc. Cusker et al. 2008)- Sig. G promoter (DBTBS)-Sig. E promoter (DBTBS)- Sle. B CDS http: //www. ncbi. nlm. nih. gov/nuccore/1146195? from=12631&to=13548&report=gbwithparts – Cwl. D CDS http: //www. ncbi. nlm. nih. gov/nuccore/1177247? from=567&to=1280&report=gbwithparts. 10 RBS (BBa_K 090505)- rcs. A CDS (BBa_K 137113)- ter 11 Sig. A- RBS- Lux. I (NCBI)-ter 12 Sig. A- RBS- Las. I (NCBI)-ter

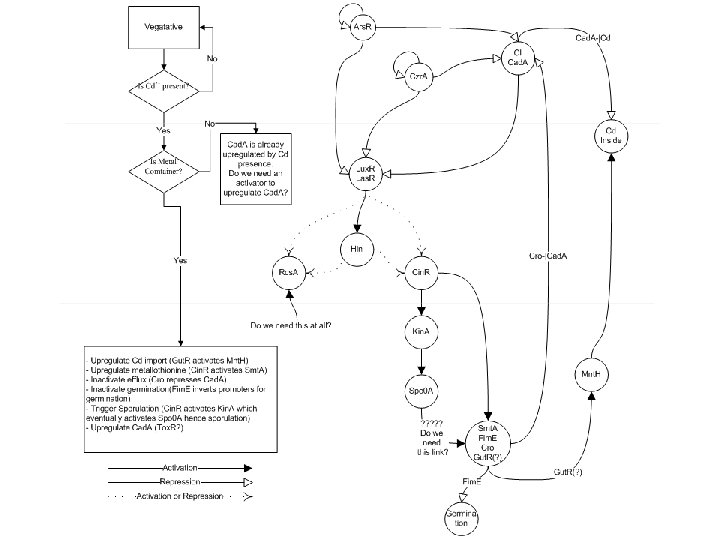
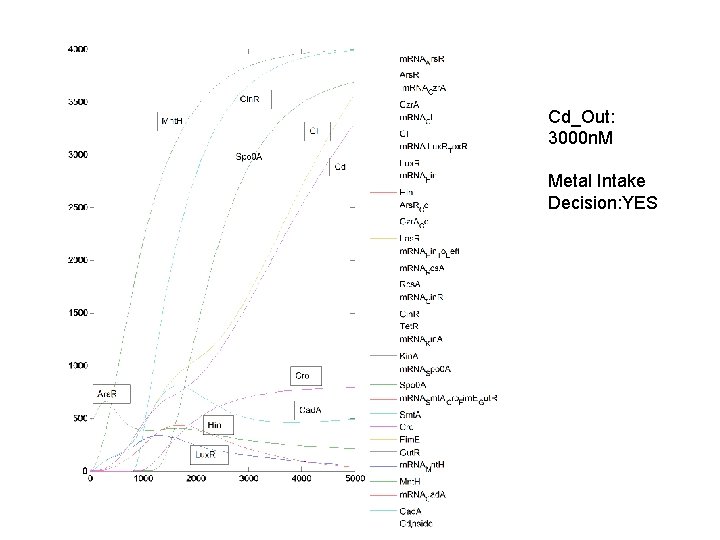
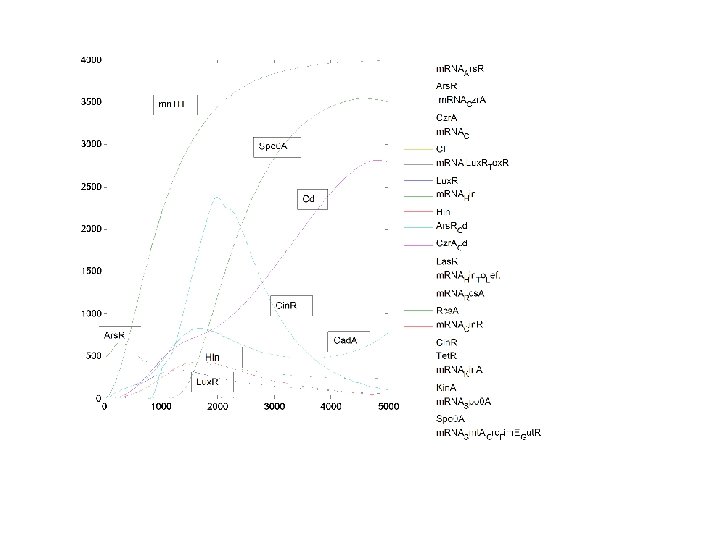
Cd_Out: 3000 n. M Metal Intake Decision: YES



Cd_Out: 3000 n. M Metal Intake Decision: NO





Lab work: what we hope to prove 1. Need to show that the sequence flips in the presence of cadmium. We have added GFP and RFP expression to the left and right sides respectively so we can see when the sequence has flipped. 2. Need to show that there is a biased heads or tails effect happening- GFP should be expressed but a lot less than RFP in the presence of cadmium. 3. Need to show that the sequence doesn’t flip in the absence of cadmium 4. Need to show that we can trigger sporulation using the switch (Kin. A). 5. Need to show that we can prevent germination in the presence of cadmium (Fim. E) 6. Need to show that in the presence of cadmium the Fim. E invertase flips the promoter for the germination genes and this is why there is no germination (YFP). 7. Need to show that metallothionein will be located to the spore coat. 8. Test the switch without using the Cd in the lab. (By adding autoinducers of Lux. R and Las. R)

Questions 1. We didn’t use the activator on the left hand side of the switch, as we think it won’t work due to decay of the proteins before they are needed. If they wouldn’t decay when they are in the spore then this would work. 2. Do we need a link between Spo 0 A and the metal container decision proteins? 3. How do we link sensing cadmium to the adjustment of sporulation? 4. When the metal container decision is ‘No’ we haven’t expressed a protein that will upregulate Cd efflux as we think this will happen in the cell anyway. As an alternative we could express a transcription factor that will repress Ars. R and Czr. A expression or an activator that can upregulate Cad. A (Tox. R? ) 5. Where to place the RE sequence? 6. Why are we choosing Hix. C of Wild type Hix. L/R (Hix. C is 16 fold slower than wt) (Davidson Bio. Brick- BBa_J 44000 Hix. C) 7. Can we use cadmium in the Lab- otherwise to test our system we need another external control mechanism. Perhaps IPTG-Lac. I-Tet. R- Czr. A/Ars. R orinduce the activators on the left and right hand side of the switch. 8. Would that be better if we express Lux. R and Las. R constitutively and express Lux. I and Las. I upon Cd sensing? This might give us a quicker response.