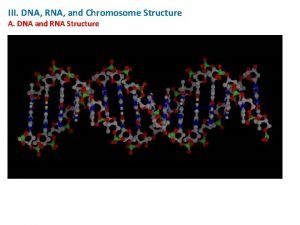
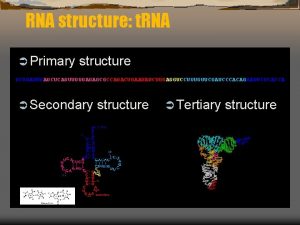
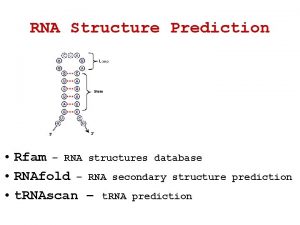
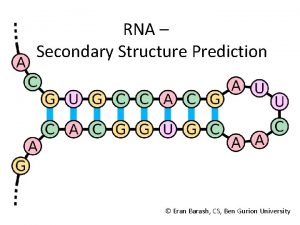
RNA Secondary Structure Prediction AAAU AAUUC UUCCG CCGG












- Slides: 46

RNA Secondary Structure Prediction AAAU AAUUC UUCCG CCGG G G Karen M. Pickard CISC 889 Spring 2002

Outline • Review: RNA v DNA • Predicting RNA secondary structure • Features of RNA secondary structure • Assumptions • Methods of RNA structure prediction CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

AAA AAAU AAUUC UUCCG CCGG G G Review • What is RNA • RNA vs. DNA CISC 889 Spring 2002

• RNA is ribonucleic acid, closely related • • to deoxyribonucleic acid or DNA. RNA is the only biological polymer that serves as both a catalyst (like proteins) and as information storage (like DNA). RNA is structurally very similar to DNA with three main differences: CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

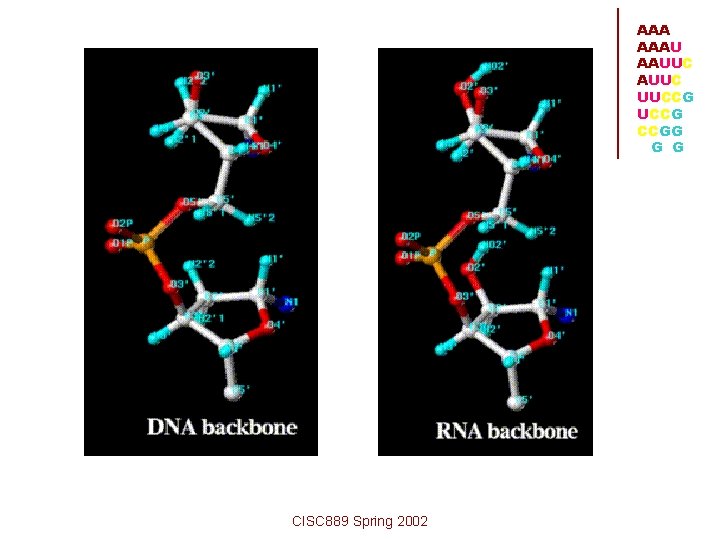
AAA AAAU AAUUC UUCCG CCGG G G • The DNA base Thymine is replaced by • • Uracil in RNA. This makes the RNA alphabet A, C, G, U rather than the DNA alphabet A, C, G, T. The phosphate sugar backbone of RNA is built out of ribose instead of deoxyribose. RNA is synthesized as a single stranded molecule. CISC 889 Spring 2002

AAA AAAU AAUUC UUCCG CCGG G G CISC 889 Spring 2002

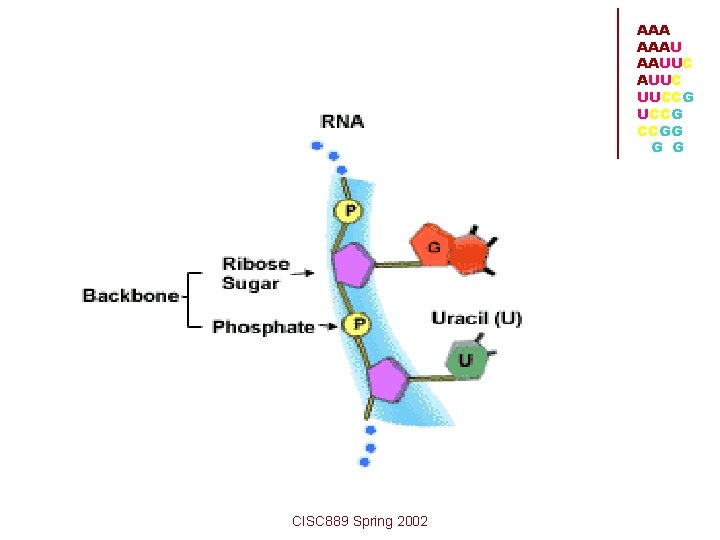
• Each ribonucleotide contains • a phosphate group • a sugar group (ribose) • a base • The polymer is formed by the linkage of the phosphate groups. The non-planar 5 member ribose ring connects the phosphate to the base. Finally, the bases are connected to the ribose group. Only the bases differ. CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

AAA AAAU AAUUC UUCCG CCGG G G CISC 889 Spring 2002

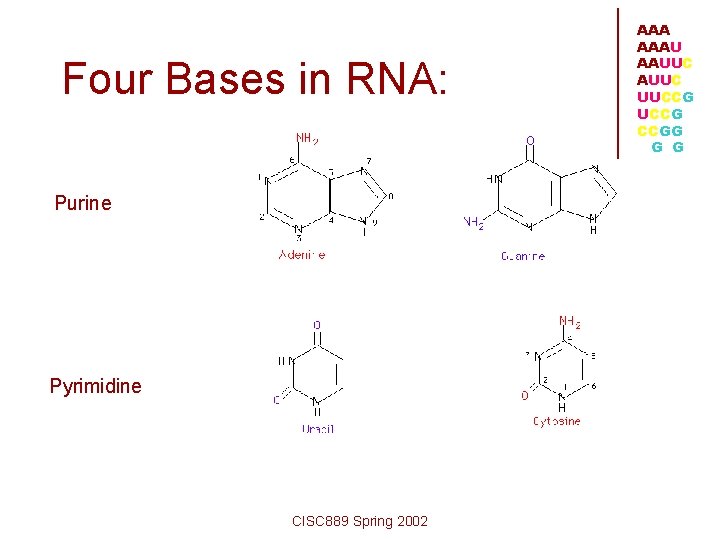
Four Bases in RNA: Purine Pyrimidine CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

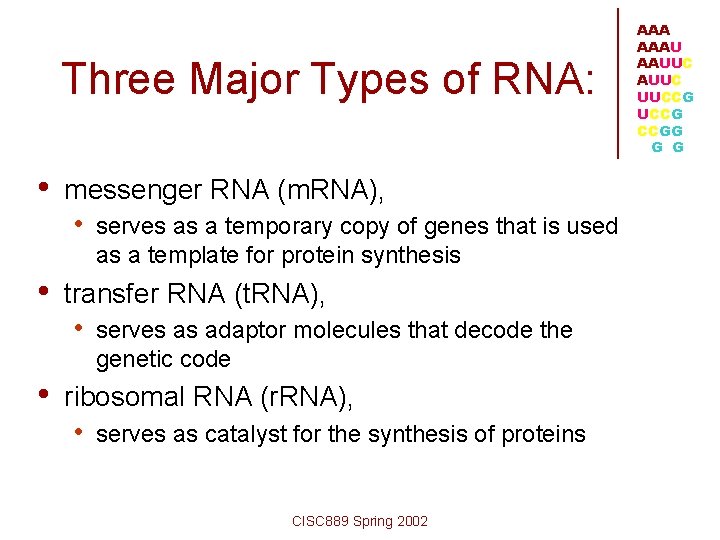
Three Major Types of RNA: • messenger RNA (m. RNA), • serves as a temporary copy of genes that is used as a template for protein synthesis • transfer RNA (t. RNA), • serves as adaptor molecules that decode the genetic code • ribosomal RNA (r. RNA), • serves as catalyst for the synthesis of proteins CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

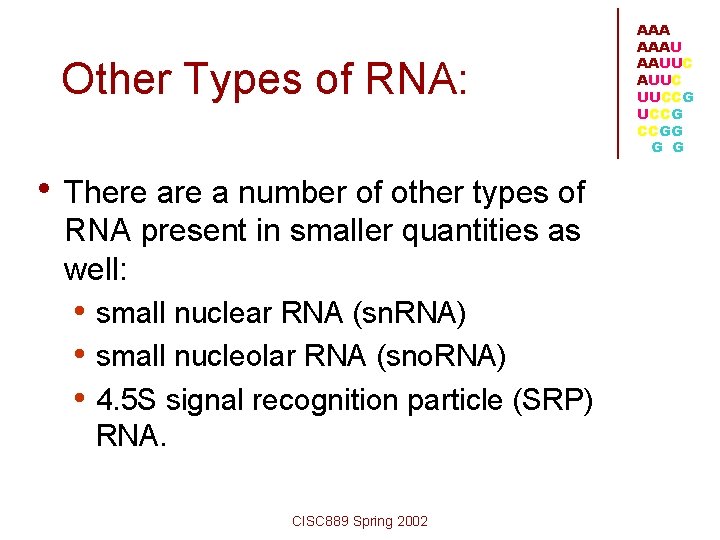
Other Types of RNA: • There a number of other types of RNA present in smaller quantities as well: • small nuclear RNA (sn. RNA) • small nucleolar RNA (sno. RNA) • 4. 5 S signal recognition particle (SRP) RNA. CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

Predicting RNA Secondary Structure • What is RNA structure prediction? • Why we study RNA structure • prediction Terminology CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

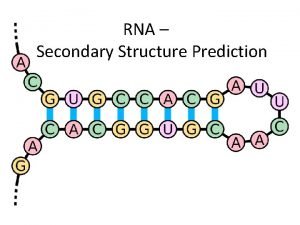
What Is RNA Secondary Structure Prediction • RNA sequence folds into functional • shape by pairing complementary bases Canonical base pairs: • complementary bases, C-G and A-U form stable base pairs • Watson-Crick pairs • G-U wobble pair CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

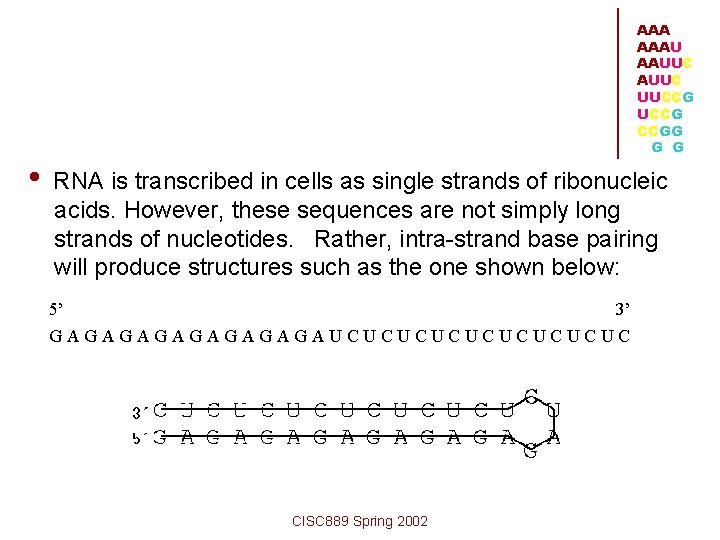
AAA AAAU AAUUC UUCCG CCGG G G • RNA is transcribed in cells as single strands of ribonucleic acids. However, these sequences are not simply long strands of nucleotides. Rather, intra-strand base pairing will produce structures such as the one shown below: 5’ 3’ GAGAGAGAUCUCUCUCUC CISC 889 Spring 2002

Why Study RNA Structure Prediction • Secondary structure related to function • Drug, viral research CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

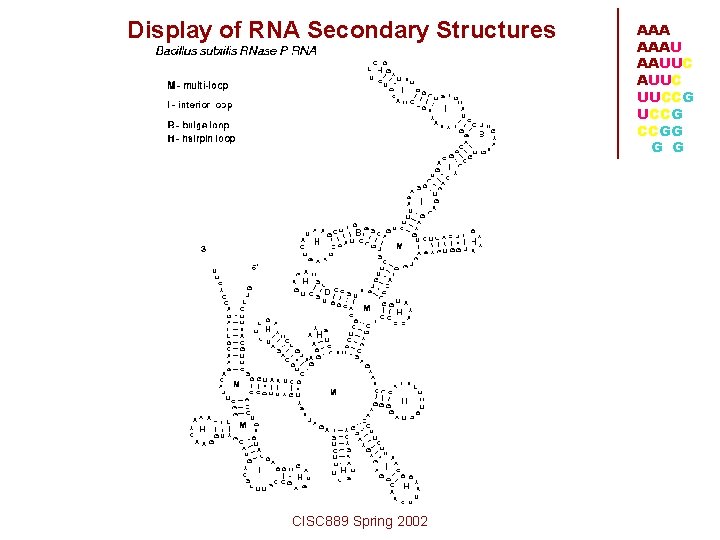
Terminology of RNA Secondary Structures • Stacked pairs • Base-pairing/base-stacking interactions. Usually Watson-Crick pairs, but G-Us are not uncommon, and other noncanonical pairs do occur • Hairpin Loop • Bulge Loop • Interior Loop CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

AAA AAAU AAUUC UUCCG CCGG G G • Multi-loop • Pseudoknots • often not considered to be "secondary structure“--most RNA secondary structure algorithms ignore them CISC 889 Spring 2002

Display of RNA Secondary Structures CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

Features of RNA Secondary Structure: • Intermediate step to 3 -D structure • Double-stranded regions formed by • single-stranded molecule folding back on itself Downstream run of bases must be complementary to upstream run of bases so Watson-Crick pairing can occur CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

Features (Continued) • G-C base pairs contribute greatest • • energy stability. A-U base pairs contribute less stability. G-U base pairs (wobble pairs) contribute least stability. CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

Assumptions: AAAU AAUUC UUCCG CCGG G G • That the most likely structure is similar to • • the most energetically stable structure. The energy associated with a particular base pair in a double-stranded region is influenced only by the previous base pair. The structure is to be formed in a manner that does not produce any knots. CISC 889 Spring 2002

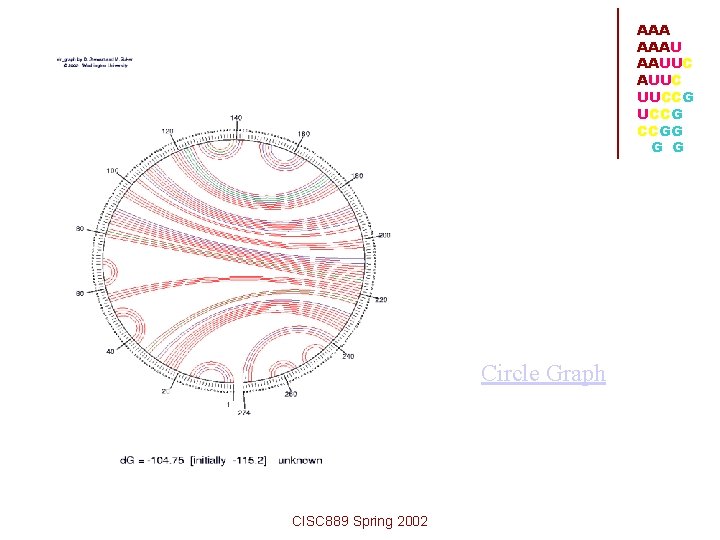
Representation of Assumptions: • Draw sequence in circular form • Paired bases joined by arcs • If structure to be free of knots, none of • the arcs must cross (If lines cross, psuedoknot) CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

AAA AAAU AAUUC UUCCG CCGG G G Circle Graph CISC 889 Spring 2002

Methods of RNA Structure Prediction • Dot Matrix • Sequence Comparison • Minimum Free Energy • MFOLD • Covariation Analysis of RNA Sequences • Context-Free Grammars CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

Dot Matrix Sequence Comparison • Find stretches of self-complementary • • regions Visual representation is easy to evaluate No need to compensate for gaps CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

AAA AAAU AAUUC UUCCG CCGG G G CISC 889 Spring 2002

Weaknesses: • No "score“, evaluation produced • For long sequences, too slow for a • database search Doesn't actually align sequences CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

Minimum Free Energy • Every base compared to every other • • • base, similar to dot matrix analysis Diagonal indicates potential doublestranded area Sum negative base-stacking energies for each pair bases in diagonal Add estimated positive energies for loops CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

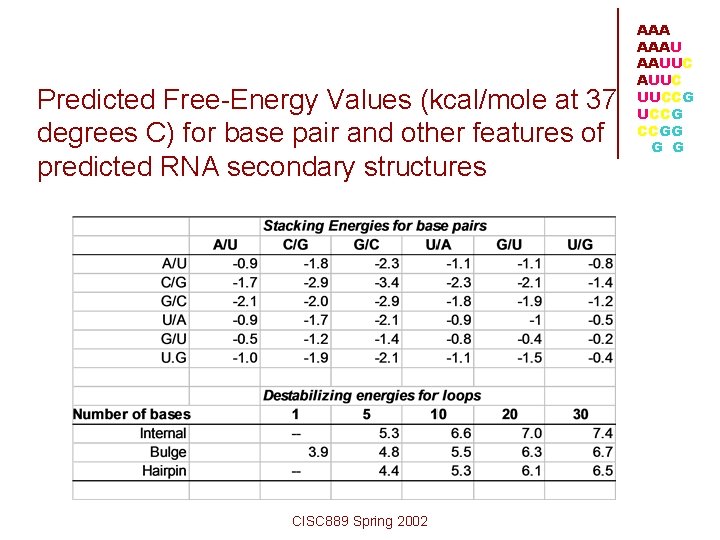
Predicted Free-Energy Values (kcal/mole at 37 degrees C) for base pair and other features of predicted RNA secondary structures CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

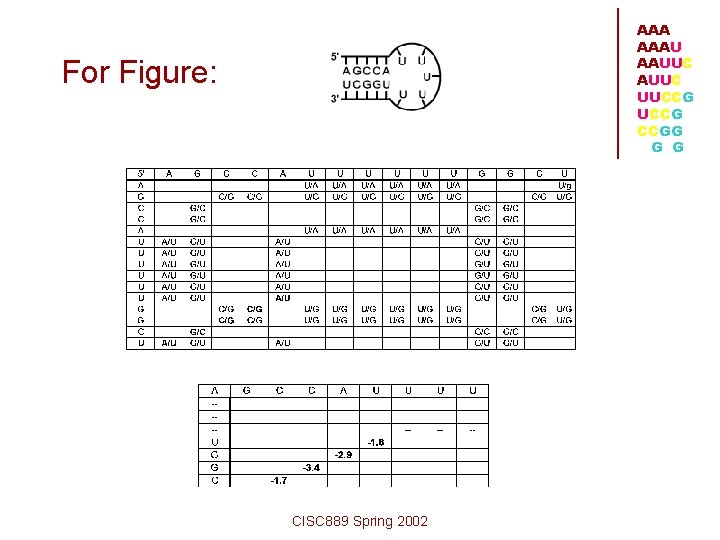
AAA AAAU AAUUC UUCCG CCGG G G For Figure: CISC 889 Spring 2002

A/U followed by G/C = -1. 7 kcal/mol G/C followed by C/G = -3. 4 kcal/mol C/G followed by C/G = -2. 9 kcal/mol C/G followed by A/U = -1. 8 kcal/mol sum = -9. 8 kcal/mol In the structure, there are five U's in the hairpin loop region, which has a predicted destabilizing energy of +4. 4 kcal/mol. Therefore, the total free energy reduction predicted for the molecule given the above secondary structure is: -9. 8 kcal/mol + 4. 4 kcal/mol = -5. 4 kcal/mol • Free Energy Calculation CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

MFOLD (Michael Zucker) • Predicts non-based-paired interactions • Predicts several structures having • energies close to minimum free energy Accurately depict structures of RNA molecules derived from comparative sequence analysis CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

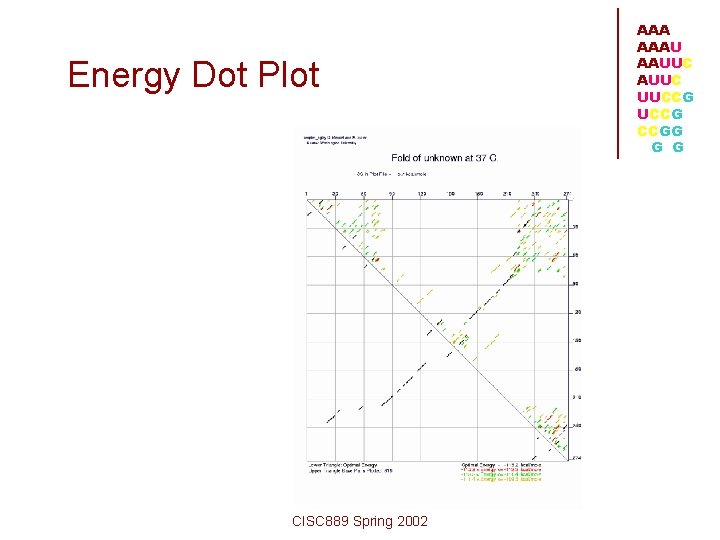
Energy Dot Plot CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

Representations of MFOLD • Display Structure • Most widely used. Closest to physical structure CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

Limitations of MFOLD • Does not compute all structures within a given energy range of the minimum free -energy structure. • No alternative structures are produced that • have the absence of base pairs in the best structure If two sub-structures are joined by a stretch of unpaired bases, no structures are produced that are suboptimal for both CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

Covariation Analysis of RNA Sequences • Based on information theory applied to • • AAAU AAUUC UUCCG CCGG G G biology After transcription, RNA spliced by cell Splicing done at donor and receptor sites Height of stack shows degree of conservation—proportional to frequency in acceptors RNA Structure Logo CISC 889 Spring 2002

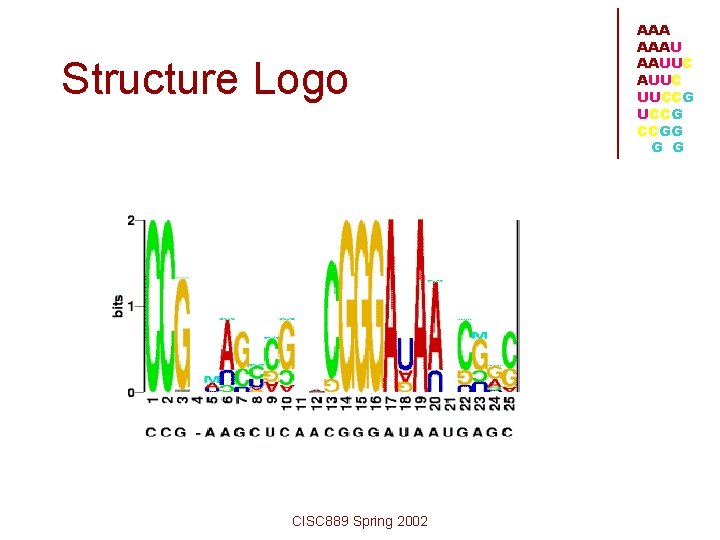
Structure Logo CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

AAA AAAU AAUUC UUCCG CCGG G G • Needs multiple alignment of a family of RNA • to identify columns of high information contents These columns indicate conservation which is essential in secondary structure and base pairing CISC 889 Spring 2002

Stochastic Context Free Grammars (SCFG) • Each RNA structure can be specified by • • a stochastic context-free grammar like that for a programming language This can be used to describe and classify RNAs Models can be used to search the DNA genome for RNA genes. CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

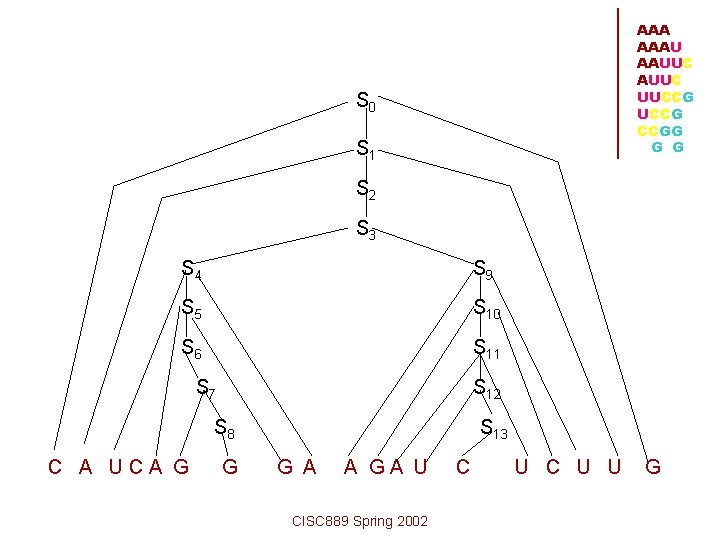
AAA AAAU AAUUC UUCCG CCGG G G For sequence CAUCAGGGAAGAUCUCUUG Productions P = { S 0 S 1, S 1 C S 2 G, S 2 A S 3 U, S 3 S 4 S 9, S 4 U S 5 A, S 7 G S 8, S 8 G, S 9 A S 10 U, S 10 G S 11 C, S 11 A S 12 U, S 5 C S 6 G, S 6 A S 7 S 12 U S 13, S 13 C CISC 889 Spring 2002 }

Derivation S 0 S 1 CS 2 G CAS 3 UG CAS 4 S 9 UG CAUS 5 AS 9 UG CAUCS 6 GAS 9 UG CAUCAS 7 GAS 9 UG CAUCAGS 8 GAS 9 UG CAUCAGGGAAS 10 UUG CAUCAGGGAAGS 11 CUUG CAUCAGGGAAGAUS 13 UCUUG CAUCAGGGAAGAUCUCUUG CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G

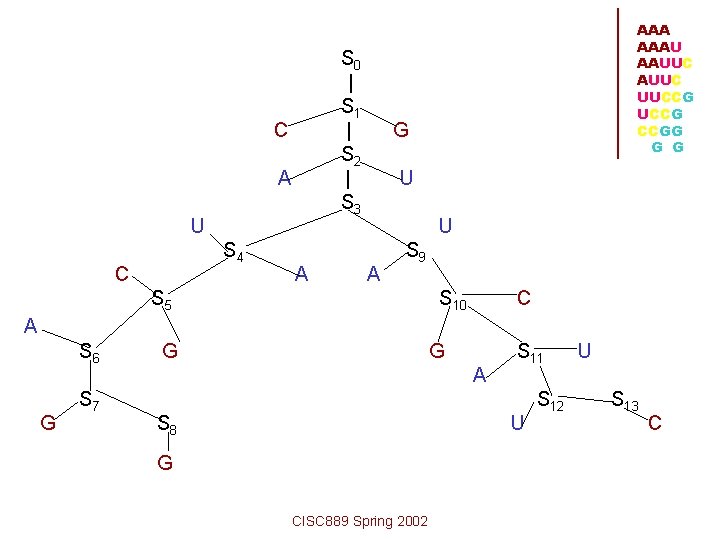
AAA AAAU AAUUC UUCCG CCGG G G S 0 S 1 S 2 S 3 S 4 S 9 S 5 S 10 S 6 S 11 S 7 S 12 S 8 C A UCA G G S 13 G A A GA U CISC 889 Spring 2002 C U U G

S 0 | S 1 | S 2 | S 3 C A U S 4 C A AAAU AAUUC UUCCG CCGG G U U A S 9 S 5 S 10 C A S 6 G S 7 G G A S 8 S 11 U G CISC 889 Spring 2002 S 12 U S 13 C

AAA AAAU AAUUC UUCCG CCGG G G Summary • Methods include • Analysis of all possible combinations of • potential double-stranded regions by energy minimization methods Identification of base covariation that maintains secondary (and tertiary) structure of RNA during evolution CISC 889 Spring 2002

AAA AAAU AAUUC UUCCG CCGG G G References • • • Mount, Daniel W. , Bioinformatics Sequence and Genome Analysis, Cold Spring Harbor Laboratory Press, 2001 M. Zuker, P. Stiegler (1981) Optimal computer folding of large RNA sequences using thermodynamic and auxiliary information, Nucl Acid Res 9: 133 -148 J. Gorodkin, L. J. Heyer, S. Brunak and G. D. Stormo. Displaying the information contents of structural RNA alignments: the structure logos. Comput. Appl. Biosci. , Vol. 13, no. 6 pp 583 -586, 1997. T. D. Schneider and R. M. Stephens. Sequence logos: a new way to display consensus sequences. Nucleic Acids Research, Vol. 18, no. 20, pp 6097 -6100, 1990. Yizong Cheng, University of Cincinnati, RNA Secondary Structure, cheng. ececs. uc. edu Chen, R. O. , Felciano, R. , and Altman, R. B. 1997. RIBOWEB: Linking structural computation to a Knowledge Base of published experimental data, Ismb 5: 84 – 87. CISC 889 Spring 2002

RNA Structure Databases • Michael Zucker's MFOLD • RNABase: The RNA Structure • Database RNA databases CISC 889 Spring 2002 AAAU AAUUC UUCCG CCGG G G
 Rna secondary structure prediction
Rna secondary structure prediction Rna secondary structure prediction
Rna secondary structure prediction Phd secondary structure prediction
Phd secondary structure prediction Canonical base
Canonical base Nussinov jacobson algorithm
Nussinov jacobson algorithm Pseudoknot structure
Pseudoknot structure 3 componets of dna
3 componets of dna Rna in chromosomes
Rna in chromosomes Jalview
Jalview To make predictions we often use
To make predictions we often use Championship branch prediction
Championship branch prediction Corner prediction
Corner prediction Chapter 26 hunger games
Chapter 26 hunger games Variance in regression
Variance in regression Merit prediction
Merit prediction Penggorengan keripik
Penggorengan keripik Make a prediction about kenny and franchesca
Make a prediction about kenny and franchesca Good readers make prediction by
Good readers make prediction by Make a prediction
Make a prediction Fb24 prediction
Fb24 prediction Proyeksi berdasarkan kedudukan sumbu simetri
Proyeksi berdasarkan kedudukan sumbu simetri Hypothesis vs prediction
Hypothesis vs prediction Branch prediction
Branch prediction On formalizing fairness in prediction with machine learning
On formalizing fairness in prediction with machine learning Masta prediction
Masta prediction Branch prediction in computer architecture
Branch prediction in computer architecture Branch prediction in computer architecture
Branch prediction in computer architecture Branch prediction
Branch prediction Punnett square for brown and green eyes
Punnett square for brown and green eyes Residual standard deviation
Residual standard deviation Integer pipeline stages of pentium processor
Integer pipeline stages of pentium processor Coefficient of determination formula in regression
Coefficient of determination formula in regression Prediction%20cfa
Prediction%20cfa What is branch prediction logic
What is branch prediction logic Tournament branch predictor
Tournament branch predictor Mathematical models for impact prediction
Mathematical models for impact prediction Prediction error method
Prediction error method Branch prediction in computer architecture
Branch prediction in computer architecture Predicting pip
Predicting pip Weisfeiler-lehman neural machine for link prediction
Weisfeiler-lehman neural machine for link prediction Peng cui tsinghua
Peng cui tsinghua What is a testable prediction
What is a testable prediction Making inferences and predictions
Making inferences and predictions Supper prediction
Supper prediction What is inferring
What is inferring Perceptual linear prediction
Perceptual linear prediction Vipers prediction questions
Vipers prediction questions