Towards RNA structure prediction 3 D motif prediction

- Slides: 17

Towards RNA structure prediction: 3 D motif prediction and knowledge -based potential functions Christian Laing Tamar Schlick’s lab Courant Institute of Mathematical Sciences Department of Chemistry New York University

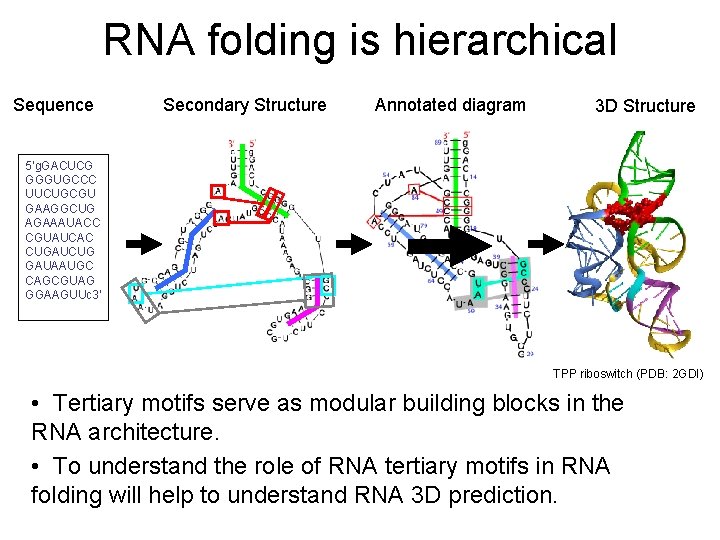
RNA folding is hierarchical Sequence Secondary Structure Annotated diagram 3 D Structure 5‘g. GACUCG GGGUGCCC UUCUGCGU GAAGGCUG AGAAAUACC CGUAUCAC CUGAUCUG GAUAAUGC CAGCGUAG GGAAGUUc 3' TPP riboswitch (PDB: 2 GDI) • Tertiary motifs serve as modular building blocks in the RNA architecture. • To understand the role of RNA tertiary motifs in RNA folding will help to understand RNA 3 D prediction.

Annotating 3 D RNA • Selected seven key RNA tertiary motifs: coaxial helix, A-minor motif, ribose zipper, tetraloop-tetraloop receptor, pseudoknot, kissing hairpin, and t. RNA D-loop: T-loop. • Searched RNA tertiary motifs via different computer programs • Annotate tertiary interaction motifs. • Perform analysis over the diagrams produced.

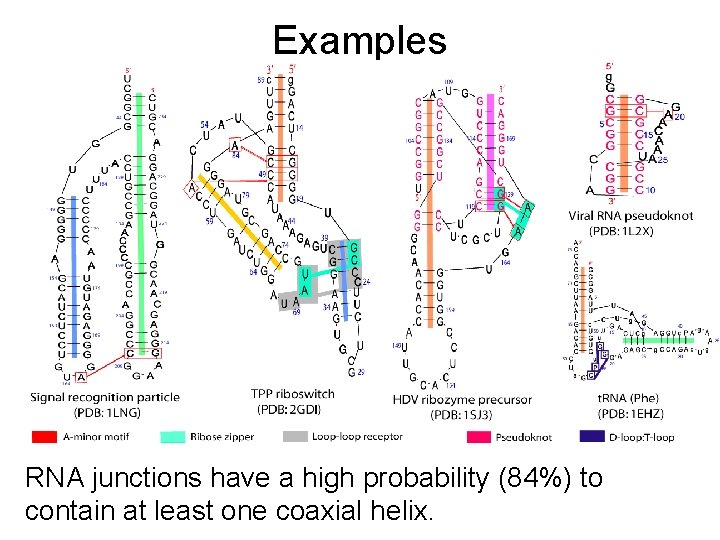
Examples RNA junctions have a high probability (84%) to contain at least one coaxial helix.

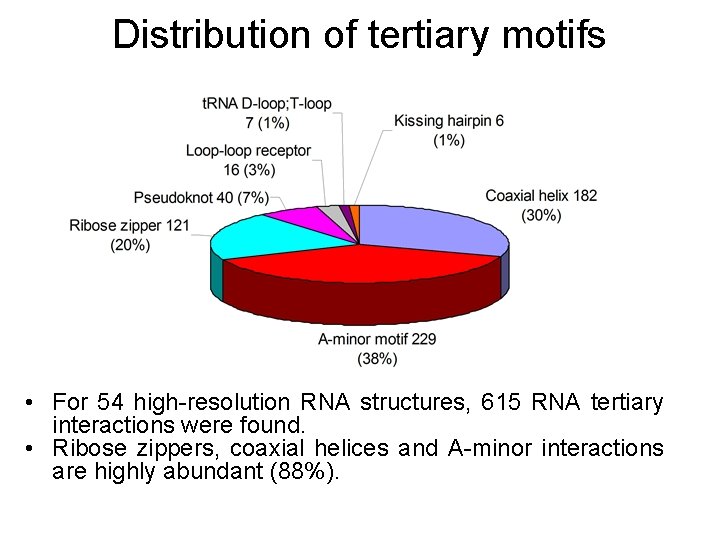
Distribution of tertiary motifs • For 54 high-resolution RNA structures, 615 RNA tertiary interactions were found. • Ribose zippers, coaxial helices and A-minor interactions are highly abundant (88%).

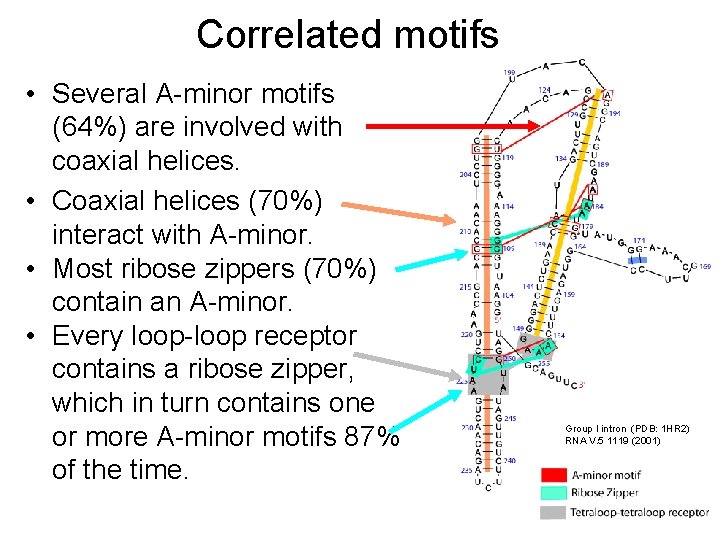
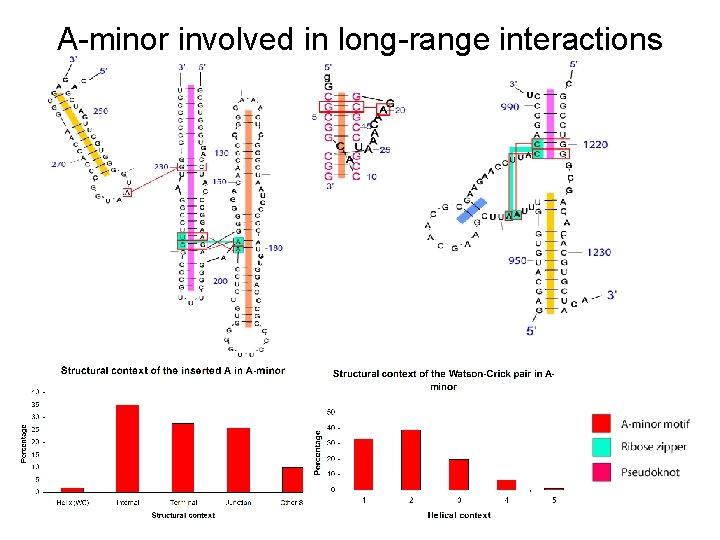
Correlated motifs • Several A-minor motifs (64%) are involved with coaxial helices. • Coaxial helices (70%) interact with A-minor. • Most ribose zippers (70%) contain an A-minor. • Every loop-loop receptor contains a ribose zipper, which in turn contains one or more A-minor motifs 87% of the time. Group I intron (PDB: 1 HR 2) RNA V. 5 1119 (2001)

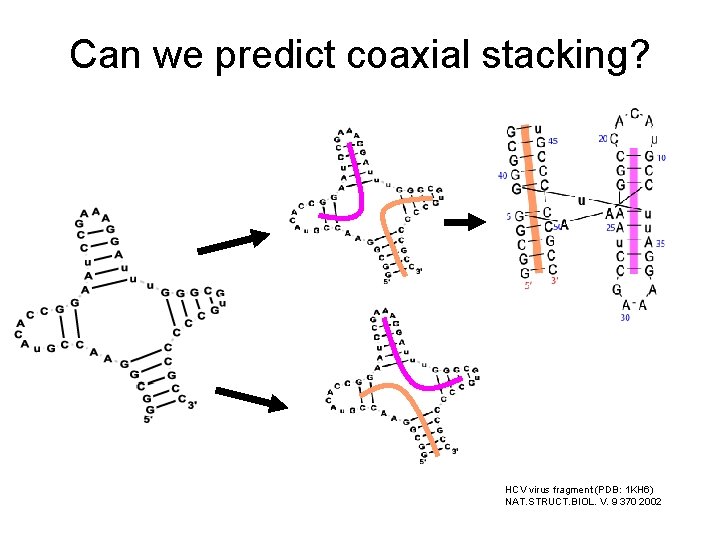
Can we predict coaxial stacking? HCV virus fragment (PDB: 1 KH 6) NAT. STRUCT. BIOL. V. 9 370 2002

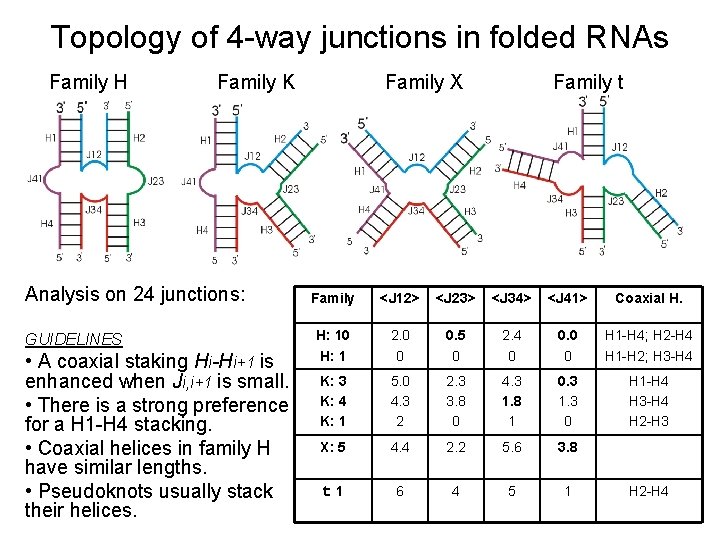
Topology of 4 -way junctions in folded RNAs Family H Family K Analysis on 24 junctions: GUIDELINES • A coaxial staking Hi-Hi+1 is enhanced when Ji, i+1 is small. • There is a strong preference for a H 1 -H 4 stacking. • Coaxial helices in family H have similar lengths. • Pseudoknots usually stack their helices. Family X Family t Family <J 12> <J 23> <J 34> <J 41> Coaxial H. H: 10 H: 1 2. 0 0 0. 5 0 2. 4 0 0 H 1 -H 4; H 2 -H 4 H 1 -H 2; H 3 -H 4 K: 3 K: 4 K: 1 5. 0 4. 3 2 2. 3 3. 8 0 4. 3 1. 8 1 0. 3 1. 3 0 H 1 -H 4 H 3 -H 4 H 2 -H 3 X: 5 4. 4 2. 2 5. 6 3. 8 t: 1 6 4 5 1 H 2 -H 4

A-minor involved in long-range interactions

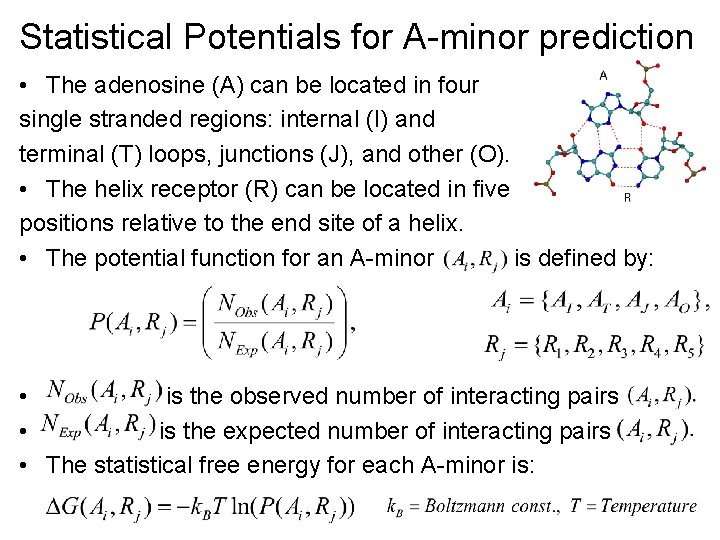
Statistical Potentials for A-minor prediction • The adenosine (A) can be located in four single stranded regions: internal (I) and terminal (T) loops, junctions (J), and other (O). • The helix receptor (R) can be located in five positions relative to the end site of a helix. • The potential function for an A-minor is defined by: • is the observed number of interacting pairs • is the expected number of interacting pairs • The statistical free energy for each A-minor is:

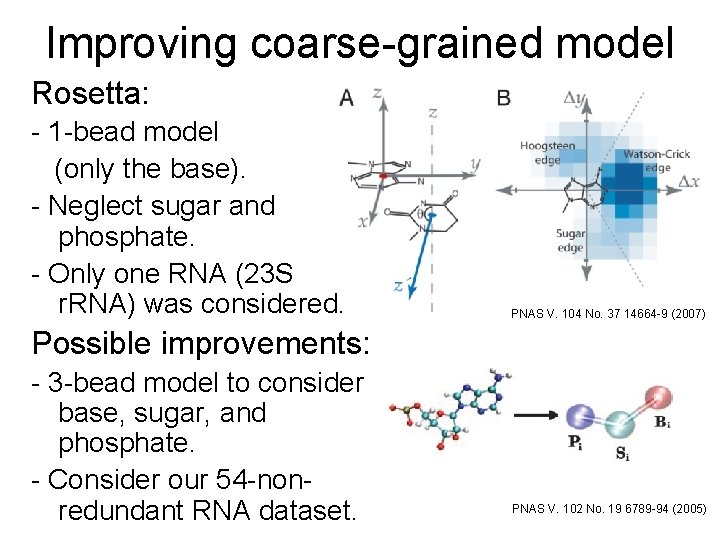
Improving coarse-grained model Rosetta: - 1 -bead model (only the base). - Neglect sugar and phosphate. - Only one RNA (23 S r. RNA) was considered. PNAS V. 104 No. 37 14664 -9 (2007) Possible improvements: - 3 -bead model to consider base, sugar, and phosphate. - Consider our 54 -nonredundant RNA dataset. PNAS V. 102 No. 19 6789 -94 (2005)

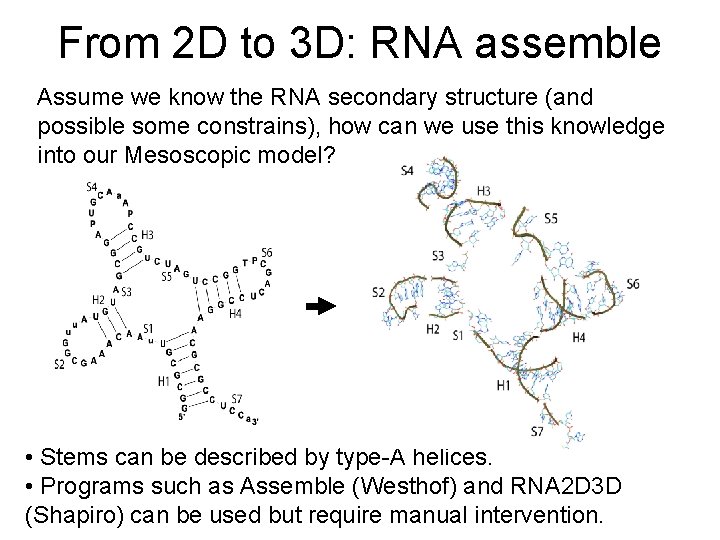
From 2 D to 3 D: RNA assemble Assume we know the RNA secondary structure (and possible some constrains), how can we use this knowledge into our Mesoscopic model? • Stems can be described by type-A helices. • Programs such as Assemble (Westhof) and RNA 2 D 3 D (Shapiro) can be used but require manual intervention.

Suggested future directions • Design statistical potentials for coaxial stacking. • The combination of A-minor/coaxial helix prediction may lead to stronger arguments. • Design statistical potentials to predict non canonical basepairs (Leontis/Westhof), and explore the possibility to use them with dynamic programming. • Test the statistical potentials (threading, decoys). • There are 272 non-redundant 4 -way junctions in the RNA junction database that can be analyzed topologically and geometrically.

Acknowledgements Yurong Xin and Tamar Schlick Many thanks to: Hin Hark Gan Sean Mc. Guffee Shereef Elmetwaly Human Frontier Science Program NSF/NIGMS initiative in Mathematical Biology (DMS 0201160)

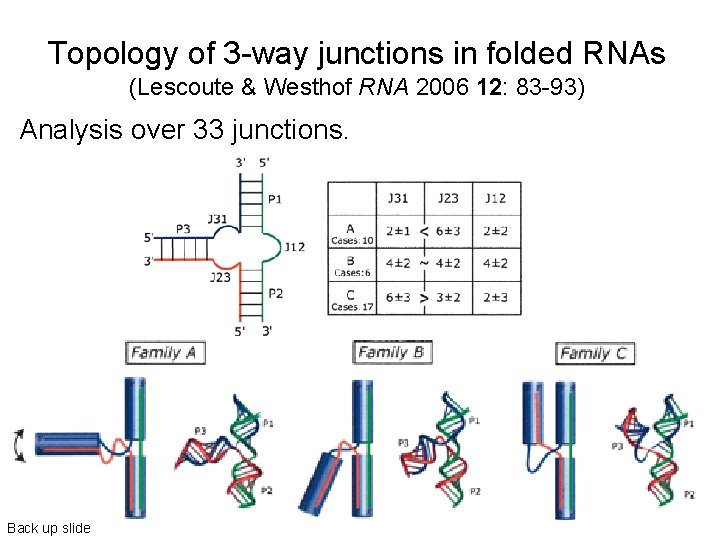
Topology of 3 -way junctions in folded RNAs (Lescoute & Westhof RNA 2006 12: 83 -93) Analysis over 33 junctions. Back up slide

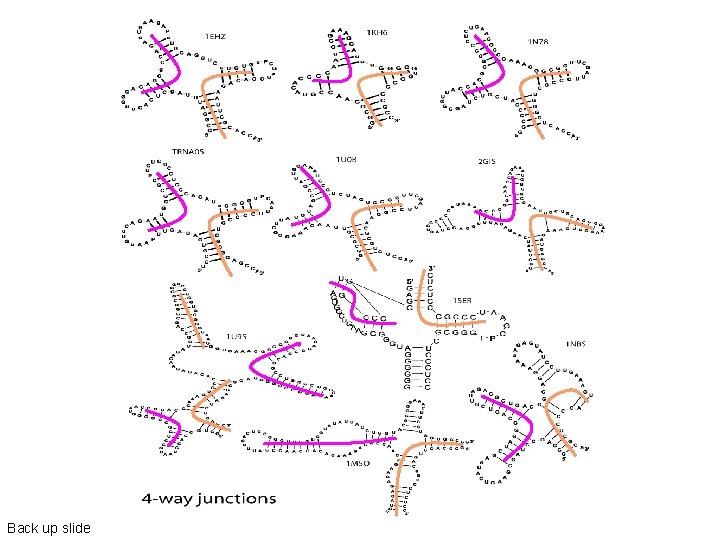
Back up slide

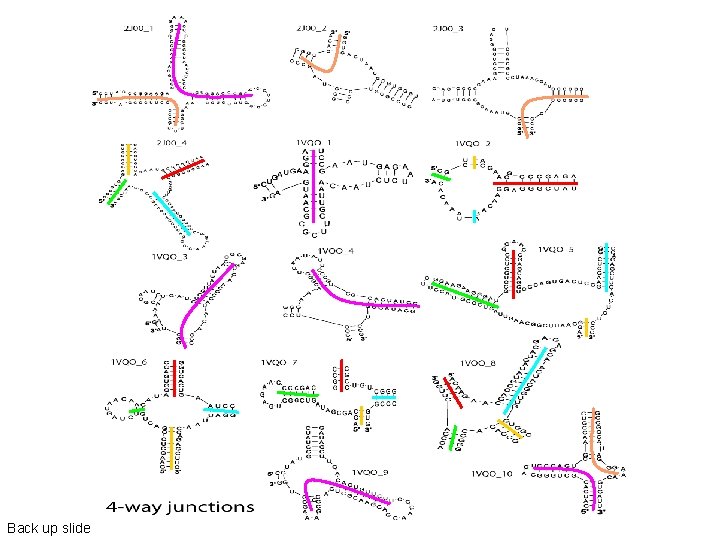
Back up slide